Fig. 6.
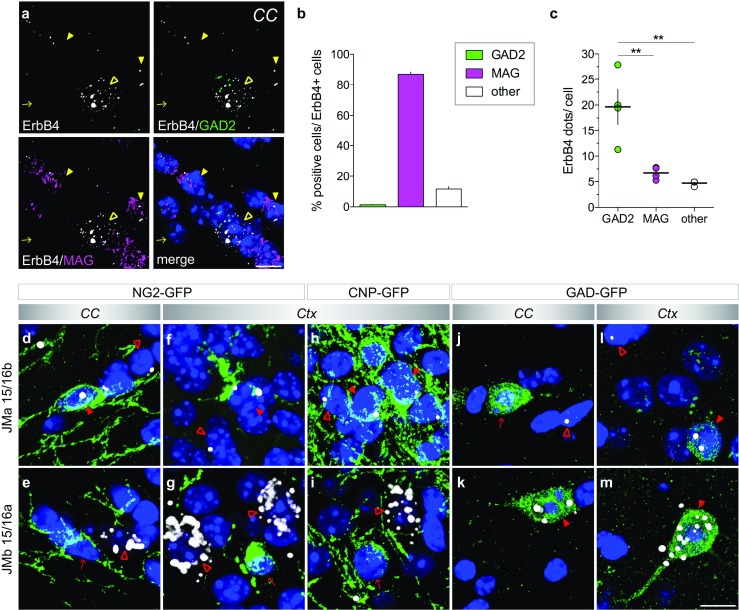
Oligodendrocytes and GABAergic neurons in the corpus callosum express different ErbB4 juxtamembrane isoforms. (a) Multiplex fluorescent in situ hybridization shows that ErbB4 (white) is expressed in both GAD2-positive GABAergic neurons (green; open yellow arrowheads) and MAG-positive oligodendrocytes (magenta; yellow arrowheads) in the corpus callosum (arrow ErbB4-negative cell). Note that dots are smaller compared to single-pair probe ISH, as signals are not enzymatically amplified. (b, c) Quantification of data shown in A (n = 4). (b) The majority of ErbB4+ cells in the corpus callosum co-expresses the oligodendrocytes marker MAG (86.95 ± 1.54%), whereas a small fraction is positive for the GABAergic marker GAD2 (1.40 ± 0.23%); 11.65 ± 1.48% of ErbB4+ cells were not labeled with either marker. (c) However, GABAergic neurons express higher levels of ErbB4 per cell than oligodendrocytes (19.65 ± 3.39 dots/cell vs. 6.73 ± 0.61 dots/cell, p = 0.0034; GAD2 vs. other 4.72 ± 0.23 dots/cell, p = 0.0013 n = 4; MAG vs. other p = 0.7614; F(2,9) = 16.53, p = 0.001; one-way ANOVA; Tukey’s multiple comparisons test: **p < 0.01). (d–m) Isoform-specific in situ hybridization using probes JMa 15/16b (d, f, h, j, l) and JMb 15/16a (e, g, i, k, m) was combined with post hoc immunohistochemistry for GFP (green) on sections from NG2-GFP (d–g), CNP-GFP (h, i) and GAD-GFP (j–m) transgenic mice. JM isoforms (white) were detected on GFP+ cells (red arrowheads), as well as on GFP negative cells (open red arrowheads) in the corpus callosum (CC) and the cortex (Ctx). Arrows depict GFP+ cells negative for JM probes. Note that the detection of JM isoforms in the corpus callosum of CNP-GFP mice was not possible because of the high density of GFP+ myelin sheaths [50]. Scale bar: 10 μm
