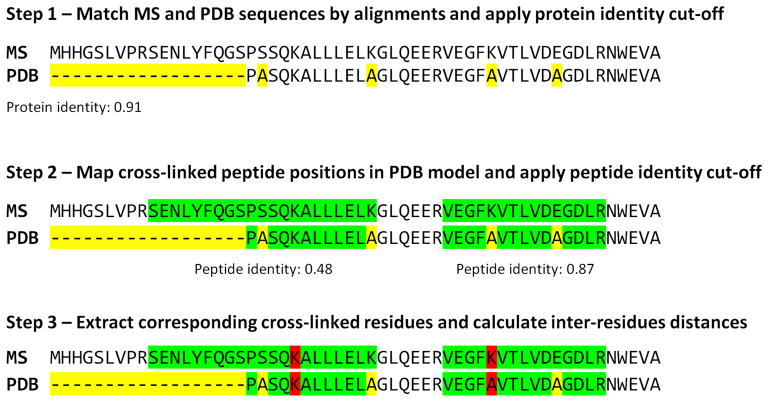
Figure 2. Algorithm for automatic mapping of cross-linked residues on PDB model.
This algorithm first finds corresponding sequences between the MS reference database and the PDB model using sequence alignment to avoid protein identifier translation issues (step 1). A user-defined protein identity cut-off is then applied and matching proteins kept. Protein sequence alignments are used as a guide to map cross-linked peptides on the PDB model to avoid issues with positional shifts, point mutations or model truncation (step 2). This algorithm allows for the use of any homologous protein models. Cross-linked residue positions are finally extracted and their Cα distance is calculated (step 3). Yellow corresponds to sequence mismatches, green to peptide sequences and red to cross-linked residues.