Figure 1.
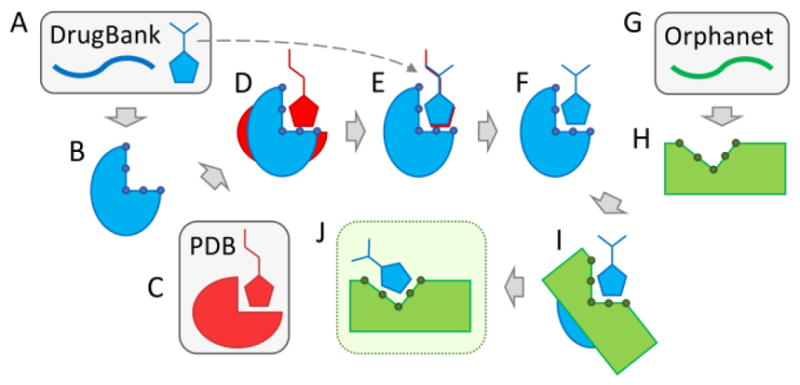
Flowchart of the drug repositioning procedure employed to construct eRepo-ORP. This protocol utilizes data from three sources, DrugBank, Protein Data Bank (PDB), and Orphanet, shown in blue, red, and green, respectively. Databases are indicated by gray boxes. (A) For a given protein sequence from DrugBank, template-based structure modeling is conducted with eThread in order to construct (B) a 3D model subsequently annotated by eFindSite with drug-binding sites and residues represented by little circles. (C) A globally similar template binding a ligand that is chemically similar to the DrugBank compound is selected from the PDB. (D) The template carrying its ligand is structurally aligned onto the DrugBank apo-structure. (E) The DrugBank compound is then aligned onto the template-bound ligand generating (F) a 3D model of the drug-target complex. (G) For a given protein sequence from Orphanet, (H) a 3D model is constructed with eThread and annotated with eFindSite. (I) A local alignment is performed for a pair of binding sites in DrugBank and Orphanet models with eMatchSite. (J) The DrugBank compound is transferred to the Orphanet model when the similarity of binding pockets in DrugBank and Orphanet models is sufficiently high and the resulting complex is refined.
