Figure 1.
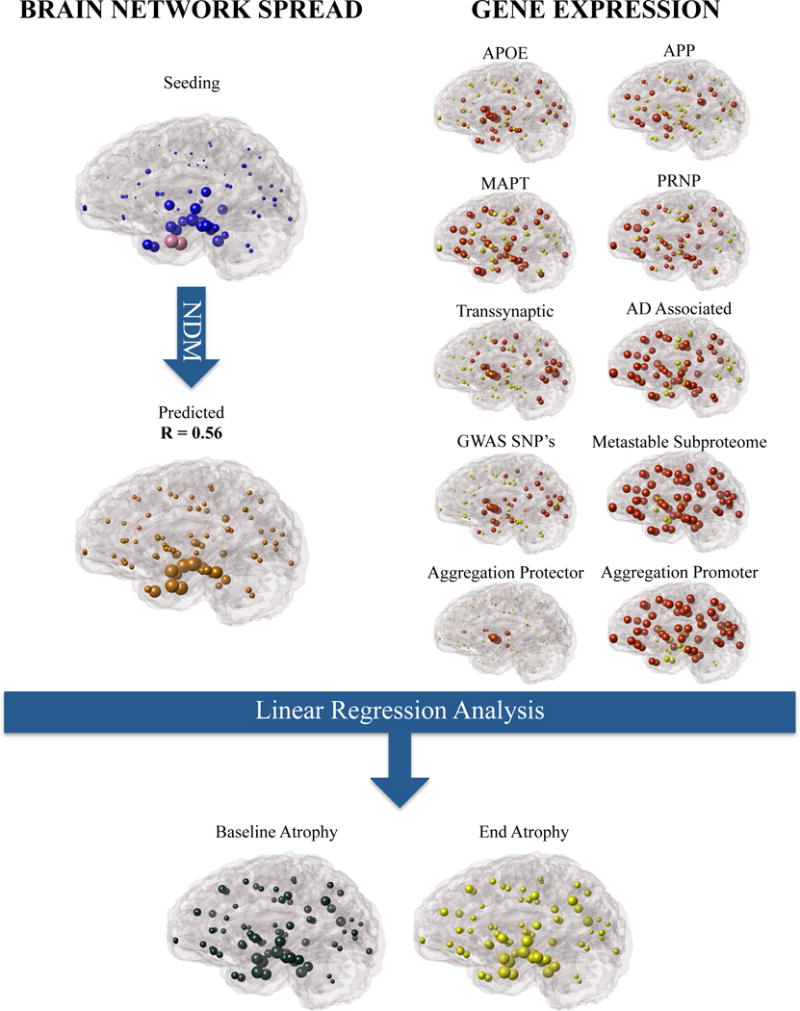
The presented methodology in which brain network connectivity models like the NDM and healthy gene expression can be used as proxies in the analysis of non-cell autonomous and cell-autonomous processes as contributors to the observed regional AD atrophy. Selected genes were compiled (AD Associated) and separated into functional subsets including (a) single genes APOE, MAPT, APP, PRNP (b) genes that were obtained from a regionally analyzed genome wide analysis (RAGWAS) (c) genes that play a role in cell to cell transmission (Transsynaptic) (d) genes that have a role in cell clearance (Metastable Subproteome) (e) genes that protect from aggregation (Aggregation Protector) and (f) genes that promote aggregation (Aggregation Promoter).
