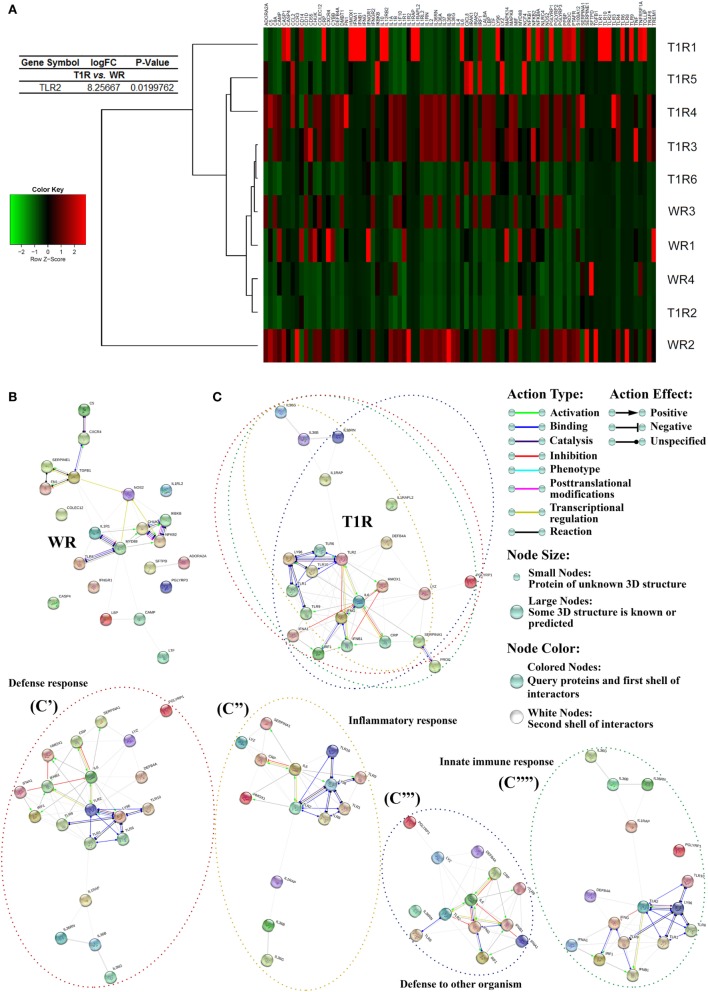
Figure 1.
Gene-expression profile of leprosy lesions showed a modulation of innate and adaptative immunity-associated genes between multibacillary patients who developed (T1R) or not (WR) reversal reactional episodes in the future. Purified mRNAs from skin lesions of multibacillary patients who developed or not reversal reaction episodes were analyzed by RT-qPCR innate and adaptative immunity array. The expression fold values of the significantly upregulated genes in WR and T1R lesions were tabulated (full data are available in Table S2 in Supplementary Material). The threshold for statistical significance was p < 0.05. (A) Heat map showing analysis of differential expression of innate and adaptative immunity-related genes in leprosy patients. Each row represents one donor. Asterisks indicate genes with differential expression. Heat map data are representative of four WR and six T1R samples. (B,C) Innate and adaptative immunity gene interaction networks in WR and T1R skin lesions. Genes with a differential expression in leprosy lesions according to autophagy PCR array analysis were visualized by STRING. The action network view. In this view, colored lines and arrow styles between genes indicate the various types of interactions. Network nodes represent genes. Edges represent gene–gene associations. (C) Interactions in genes annotated to defense response (C′), inflammatory response (C″), defense to other organism (C′″), and innate immune response (C″″) ontology terms in T1R group patients are shown. Interaction maps are representative of four WR and six T1R samples.