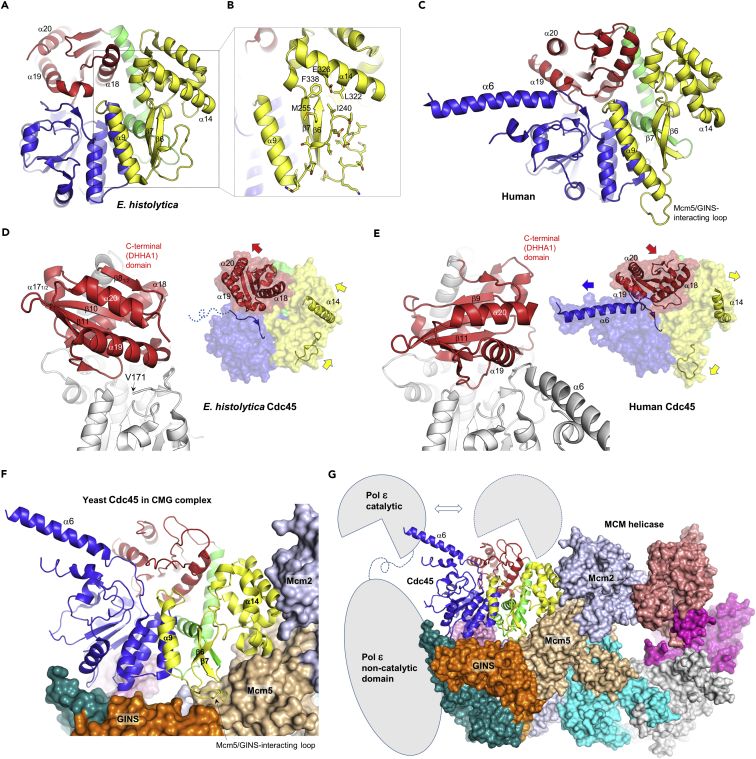
Figure 3.
Structural Comparison
(A) E. histolytica Cdc45 structure.
(B) Zoomed-in view of the region highlighted by a dotted box in panel A. The loop between α9 and β6 of E. histolytica Cdc45 takes a unique “tucked-in” conformation.
(C) Human Cdc45 structure, PDB ID: 5dgo (Simon et al., 2016).
(D) E. histolytica Cdc45, with the C-terminal DHHA1 domain highlighted in red. A possible coordination between positioning of the DHHA1 domain, α14 helix, and the Mcm5/GINS-interacting loop is depicted in the right panel.
(E) Human Cdc45 with the C-terminal DHHA1 domain highlighted. Note distinct positioning of the DHHA1 domain, bent α14 helix, and untucked Mcm5/GINS-interacting loop.
(F) Yeast CMG helicase cryo-EM structure (PDB ID: 3jc6) (Yuan et al., 2016) viewed from the N-terminal face of MCM, showing extensive interactions between Cdc45 (ribbon) and Mcm2/5 and the GINS subunits (surface).
(G) A full view of the CMG helicase, with the spatial relationship between helicase and DNA polymerase ɛ shown schematically. The catalytic domain of DNA polymerase ɛ can take alternative positions, one of which is stabilized by interaction with the tip of α6 helix of Cdc45 (Zhou et al., 2017).