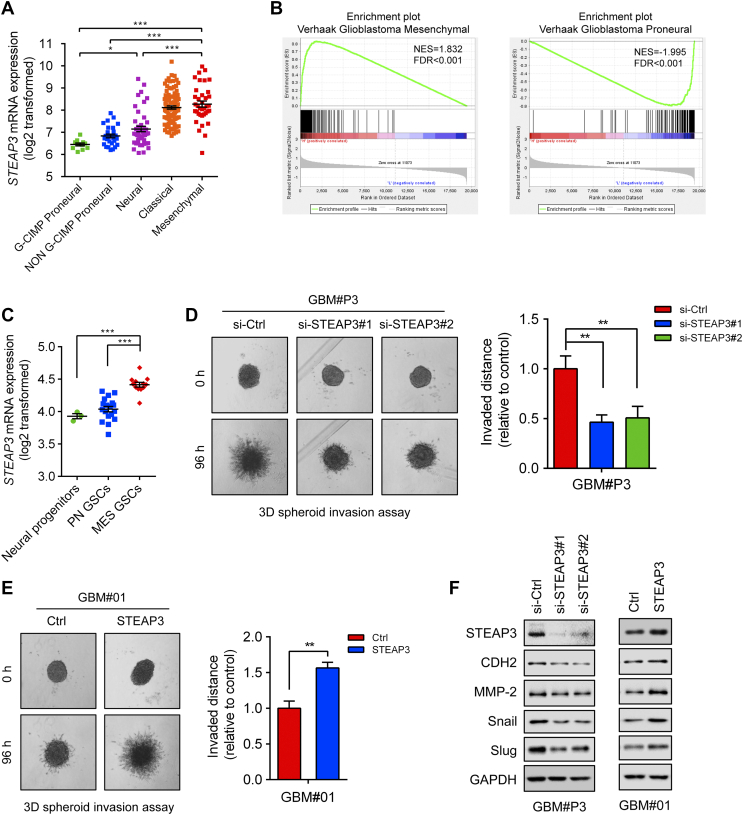
Figure 4.
STEAP3 is enriched in the GBM mesenchymal molecular subtype and promotes cell invasive potential. (A) STEAP3 mRNA expression in different molecular subtypes (CpG island methylator phenotype (G-CIMP) proneural, non–G-CIMP proneural, neural, classical, mesenchymal) from the TCGA GBM microarray dataset. (B) GSEA enrichment analysis of mesenchymal and proneural signatures (Verhaak) in STEAP3highvs STEAP3low samples in the TCGA GBM dataset. Normalized enrichment score (NES) and FDR are shown for each plot. (C) STEAP3 mRNA expression in neural progenitor cells (n = 3), PN GSCs (n = 18), and MES GSCs (n = 12) in microarray data. (D) Representative images of spheroids in 3D invasion assay for GBM#P3 GSCs transfected with si-Ctrl and si-STEAP3 and evaluated at 0 h and 96 h. The distance of invading cells from the tumor spheres was determined after 96 h. (E) 3D invasion assay at 96 h for GBM#01 GSCs overexpressing STEAP3. (F) Western blot for protein levels of key factors involved in mesenchymal transition in lysates (20 μg) prepared from GBM#P3 and GBM#01 GSCs. GAPDH was used as a loading control. Data are shown as the mean ± SEM from three independent experiments. *P < .05; **P < .01; ***P < .001.