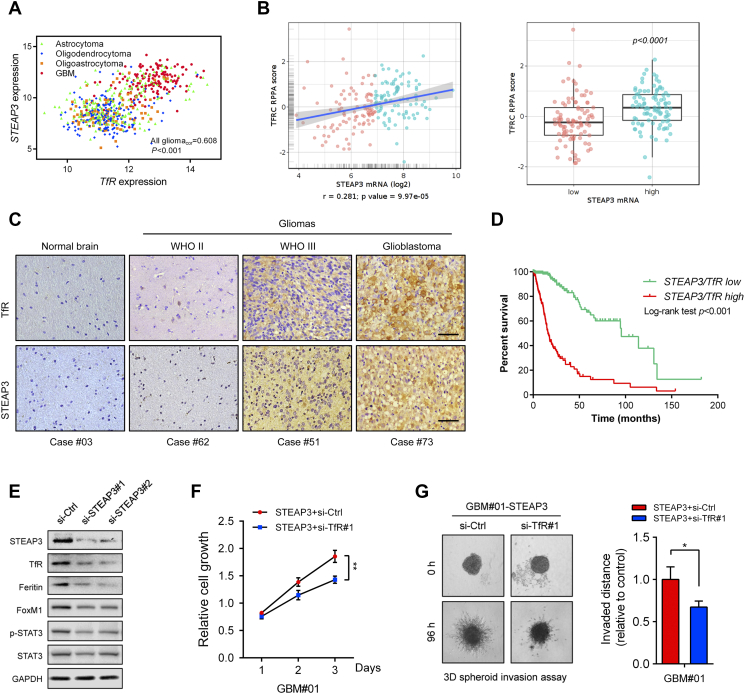
Figure 6.
STEAP3 activates the TfR-STAT3 pathway in GBM cells in vitro. (A) Correlation between STEAP3 and TfR mRNA expression in gliomas from the TCGA dataset. The statistical significance of correlation was evaluated using a linear regression model (TCGA all gliomacor = 0.608, P < .001). (B) Correlation between STEAP3 mRNA and TfR protein levels in TCGA GBM patients (r = 0.281, P = 9.97e-05). TfR protein levels were increased in the STEAP3high group (P < .001). (C) Representative images of paired IHC staining for TfR and STEAP3 in individual cases of normal brain and different pathological grades of astrocytoma. Scale bar = 100 μm. (D) Kaplan–Meier curve analysis of survival data from patients based on low/high co-expression of STEAP3/TfR. P-values were obtained from the log-rank test. (E) Western blot analysis of TfR, Ferritin, FoxM1, STAT3 and phosphorylated-STAT3 in GBM#P3 GSCs transfected with si-Ctrl and si-STEAP3. GAPDH was used as a loading control. (F) Growth curves generated for GBM#01-STEAP3 GSCs transfected with si-Ctrl and si-TfR in vitro. Measurements were obtained at days 1, 2, and 3 using the Luminescent Cell Viability Assay kit. (G) 3D invasion assay at 96 h for GBM#01-STEAP3 transfected with TfR knockdown siRNA. Data are shown as the mean ± SEM from three independent experiments. *P < .05; **P < .01; ***P < .001.