Abstract
Introduction:
Early identification of sepsis is critical as early treatment improves outcomes. We sought to identify threshold values of secretory phospholipase A2 (sPLA2)-IIA that predict sepsis and bacterial infection compared to nonseptic controls in an emergency department (ED) population.
Materials and Methods:
This is a prospective cohort of consenting adult patients who met two or more systemic inflammatory response syndrome (SIRS) criteria with clinical diagnosis of infectious source likely (septic patients). Controls were nonseptic consenting adults undergoing blood draw for other ED indications. Both groups had blood drawn, blind-coded, and sent to an outside laboratory for quantitative analysis of sPLA2-IIA levels. The study investigators reviewed patients’ inpatient medical record for laboratory, imaging, and microbiology results, as well as clinical course.
Results:
sPLA2-IIA levels were significantly lower in control patients as compared to septic patients (median = 0 ng/ml [interquartile range (IQR): 0–6.5] versus median = 123 ng/ml [IQR 44–507.75]; P < 0.0001). SPLA2-IIA levels were higher in patients with confirmed source (n = 28 patients, median = 186 ng/ml, 95% confidence interval = 115.1–516.8) as compared to those with no source identified or a viral source (n = 17, median = 68 ng/ml, 95% confidence interval = 38.1–122.7; P = 0.04). Using a cutoff value of 25 ng/ml, sPLA2-IIA had a sensitivity of 86.7% (confidence interval 72.5–94.5) and a specificity of 91.1% (confidence interval 77.9–97.1) in detecting sepsis.
Conclusions:
sPLA2-IIA shows potential as a biomarker distinguishing sepsis from other disease entities. Further study is warranted to identify predictive value of trends in sPLA-IIA during disease course in septic patients.
Keywords: Biomarkers, sepsis, septic shock
INTRODUCTION
Bloodstream infections or sepsis is a major cause of patient morbidity and mortality. It is estimated that sepsis affects 650,000–750,000 people each year and results in over 200,000 deaths. In fact, the incidence of sepsis has increased by about 91.3% over the last decade.[1,2] The BASES study, which looked at sepsis in hospitals in Brazil, concluded that 27% of patients admitted to the Intensive Care Unit (ICU) had severe sepsis and the mortality rate for that population was 47%.[3] According to recent data, sepsis contributes to 6% of all deaths in the US.[4] Furthermore, it comes with a very high price tag: it is estimated that sepsis identification and treatment account for as much as one-quarter of all hospital charges in the US. This has been estimated to total over $24 billion a year in health-care costs.[4] When looking at emergency department (ED) patient populations, one study found that 6.4% of patients presenting to Brazil EDs had the diagnosis of severe sepsis.[5] In US EDs, suspected severe sepsis accounts for over 500,000 visits every year. Furthermore, patients presenting with severe sepsis spend an average of 5 h in the ED before admission to the hospital.[6]
Previous research suggests that in patients with septic shock, mortality rates increase by 7.6% each hour that effective antimicrobial therapy is delayed. Therefore, it is imperative that at-risk patients presenting to the ED are identified quickly and efficiently. Unfortunately, there are limited diagnostic tests available to attain this goal. Following sepsis guidelines, patients presenting with systemic inflammatory response syndrome (SIRS) criteria and a suspected source of infection should be treated for sepsis. Source of infection can be confirmed with imaging studies, urine culture, or blood culture. Currently, the gold standard for diagnosing bloodstream infections is to obtain blood cultures, typically drawn as a set of two. However, this is rarely useful acutely as sepsis can progress rapidly over the course of hours and blood cultures take days to confirm the diagnosis.[7] Blood cultures may be negative in patients who are septic from pulmonary, urinary, or soft-tissue sources. Blood culture positivity does not necessarily indicate if a patient will progress to severe sepsis or septic shock. Therefore, although blood cultures are obtained in the ED, they rarely have any effect on patient care in the critical 1st h.
Markers such as serum procalcitonin and C-reactive protein have been explored in regard to their predictive values in sepsis progression, and they have been shown to have a low sensitivity and specificity in sepsis.[8,9] Lactate levels are routinely obtained as part of a sepsis workup and are used to guide resuscitative efforts. Previous studies have shown lactate to be a predictor of mortality in septic patients, independent of vital sign abnormalities.[10] Although a lactate level may be helpful as a trend, a single level has poor prognostic value as a predictor of clinical outcomes.[11]
The secretory phospholipase A2 (sPLA2) protein family consists of 10 calcium-dependent extracellular enzymes that catalyze the hydrolysis of phospholipids at the sn-2 position, yielding proinflammatory molecules.[12] This enzyme is a part of a family of acute phase proteins which are triggered in response to inflammation. A member of this family of proteins, sPLA2-IIA, has been suspected to play a role in the innate immune response to bacterial infections. It acts by hydrolysis of the phospholipid group on the membrane lipids, leaving behind lysophosphatidylcholine (LPC). Lysis of LPC starts a cascade important in inflammation as well as in directing bactericidal activity.[13]
sPLA2-IIA levels have been shown to be elevated in animal models of sepsis as well as several preliminary human studies.[14,15,16,17] One study, looking specifically at ED populations presenting with sepsis, noted that sPLA2-IIA had a strong correlation with early sepsis diagnosis and was able to reliably distinguish the sepsis group from nonsepsis.[15] Other groups have shown that increased sPLA2-IIA plasma levels are specifically associated with patients exhibiting bacterially induced SIRS and effectively distinguish them from patients with SIRS derived from viral infections.[16,17]
In this pilot study, we used a novel ELISA assay developed and optimized by an outside laboratory to detect human sPLA2-IIA within the ng/ml range that is clinically significant as per the previous literature. We sought to identify threshold values of sPLA2-IIA that predict sepsis and bacterial infection as compared to nonseptic controls in an ED population.
MATERIALS AND METHODS
Study design
The study was a prospective cohort design of patients presenting to the ED suspected of having sepsis as compared to nonseptic controls. In septic patients, we utilized blood samples for lactate measurements, white blood cell counts (WBCs) and differentials, evidence of end-organ damage (troponin, blood urea nitrogen and creatinine, troponin values, and liver function tests), and sPLA2-IIA measurements. The patients’ electronic medical record was used for the source of clinical information, and these data were gathered by two research investigators at the clinical site. The offsite measurement of sPLA2-IIA was funded and performed by an outside facility with no access to the patients’ identity, other laboratory values, or clinical data. The participation in the study was voluntary and posed no cost, change in care, harm, or benefit to enrolled patients. Clinicians caring for the patients did not have access to sPLA2-IIA results.
Study site and population
The study site is a community-based tertiary care facility with an estimated annual ED volume of 51,000 visits and an ICU with 37 beds. The study population was derived from a convenience sample of adults presenting to the ED over the period of December 2015 to April 2016. Patients aged 18 years and older were screened for eligibility by research investigators. Patients were eligible for enrollment as study patients if they met two or more SIRS criteria with a clinical diagnosis of infectious source likely (septic patients). Control patients were also aged 18 years and older. They were identified as patients undergoing laboratory draw for reason other than sepsis (most commonly for chest pain, neurological complaints, or abdominal pain). Both control and study patients underwent written informed consent before enrollment. The study protocol was approved by the institutional review board.
Study protocol and measurements
All providers involved in enrollment of subjects underwent research training and certification as per hospital policy. After voluntary written consent, patients underwent routine blood draw procedure by nursing, medical assistant, or physician staff as per hospital protocols. The study sample (ethylenediaminetetraacetic acid plasma) was drawn with other blood tubes as indicated by the patient's clinical team. The study sample was always the last in the draw sequence and never required an additional needle stick to obtain. Blood tests obtained in septic patients are determined by hospital protocol and include complete blood count, electrolytes, urine and blood cultures, and lactate levels. Blood tests in control patients were at the discretion of the treating clinical team. These patients did not routinely have lactate levels drawn and most commonly were undergoing evaluation for atraumatic chest pain or neurologic complaints.
After collection, study blood samples were blind-coded and placed in an upright position by the nurse or physician in a designated temperature-controlled refrigerator set at 4°C. Twice weekly, collected samples were transported 50 miles to an off-site laboratory for sPLA2-IIA ELISA testing. On arrival at the offsite laboratory, each sample was centrifuged, and then plasma was removed and stored at −20°C. Once all of the study samples were collected, they were thawed and analyzed together using a sPLA2-IIA ELISA test system (ZEUS Scientific, part# ACC6301I). The sPLA2-IIA values for each sample were then provided back to the study-site investigators for decoding and evaluation against the patient-matched clinical data.
Patient data
Research associates involved in data abstraction from the clinical chart underwent training in abstraction and entered data into a standardized Excel Spreadsheet (Microsoft Excel 2007, Microsoft Corporation, Redmond, WA, USA), choosing from a closed list of possibilities. All clinical data were checked a second time by another research associate, and any discrepancies were resolved by consensus. Patient data collected from the clinical chart for septic patients included age, gender, serial lactate levels (including return to normal), ongoing vital signs (including time to resolution of SIRS), culture results, infectious source/diagnoses, and clinical outcomes. Patient data collected for control patients included chief complaint and indication for blood draw only. sPLA2-IIA levels were reported from the outside facility by blinded patient study number to clinical investigators, who then matched the levels to patient clinical data.
Definition of confirmed source and severe sepsis/septic shock
Patients were determined to have a confirmed source of their sepsis if they had positive urine cultures, blood cultures, peritoneal fluid cultures or elevated peritoneal WBCs and bands, abscesses/clinically infected wounds, or pneumonia on radiograph. Sputum cultures are not routinely obtained in patients with radiographic evidence of pneumonia with clinical symptoms, and abscesses are also not routinely cultured, so organism identification was not required to meet sepsis criteria in our study. They were determined to have severe sepsis or septic shock if they were septic and had a lactate level ≥4, an increase in their baseline creatinine of 50%, a positive troponin, or hypotension.
Data analysis
This was a pilot study with a target enrollment of 100 patients to better determine appropriate clinical threshold values for sPLA2-IIA using this assay. Data were nonparametric in distribution and were analyzed with descriptive statistics, Chi-square, and Mann–Whitney test. Data were analyzed using MedCalc (© 1993–2013, Ostend, Belgium) and VassarStats: Website for Statistical Computation (vassarstats.net, author Richard Lowry, PhD, Professor of Psychology Emeritus, Vassar College, Poughkeepsie, NY,© 1998–2013). The study was IRB approved and was supported in part by an internal grant from the study facility.
RESULTS
Demographics
One hundred patients were enrolled. Four specimens were excluded for protocol violations, five specimens were inadequate for analysis secondary to hemolysis, and one patient had an incomplete clinical record, leaving 90 for analysis. Patients are distributed as shown in Figure 1. The mean age of enrolled patients was 56.9 years (standard of deviation 17.7, range 18–92) and 51% were female. Compared to control patients, sepsis/severe sepsis patients were significantly older (mean age 62.7 vs. 51, P = 0.001) and much more likely to get admitted to the hospital (26.7% versus 97.8%, P < 0.0001). There was no difference in gender distribution.
Figure 1.
Flow diagram of patient distribution
Clinical data
Control patients were admitted for a median of 0 days (interquartile range 0–1). Twelve control patients were admitted, four for neurological complaints, two for cardiac complaints, two for gastrointestinal complaints, and one each for fall, hypotension, overdose, and global weakness. Septic patients were admitted for a median of 5 days (interquartile range 4–8.5). One patient met SIRS criteria and was believed to have an infectious source, was enrolled in the study, subsequently had improvement in his vital signs after resuscitation, and was discharged home without hospitalization with a final diagnosis of gastroenteritis. Six patients were treated in the ICU, and one died. Of 45 septic patients, 28 had confirmation of the source. The most common source was pneumonia, followed by urinary tract infections [Table 1].
Table 1.
Identified source of sepsis in study patients

Secretory phospholipase A2-IIA levels
Controlling for the presence or absence of sepsis, sPLA2-IIA levels did not vary significantly with age (P = 0.46). The median sPLA2-IIA level in controls was 0 ng/ml (interquartile range [IQR] 0–6.5), as compared to septic patients, whose median sPLA2-IIA level was 123 ng/ml (IQR 44–507.75, P < 0.0001) [Figure 2]. sPLA2-IIA levels were higher in patients in whom their sepsis had a confirmed source (n = 28 patients, median sPLA2-IIA level of 186 ng/ml, 95% confidence interval 115.1–516.8) as compared to those with no source identified or a viral source (n = 17, median sPLA2-IIA level 68 ng/ml, 95% confidence interval 38.1–122.7, P = 0.04). sPLA2-IIA levels did not, however, have discriminatory value between patients with sepsis and those with severe sepsis or septic shock (median 123 ng/ml, 95% confidence interval 67.3–180.6 as compared to median 144 ng/ml, 95% confidence interval 33.4–525, P = 0.79). sPLA2-IIA levels had a weak relationship with lactate levels for 43 patients who had lactate levels drawn (r2 = 0.11, P = 0.03). sPLA2-IIA also had a weak relationship with hospital length of stay (r2 = 0.11, P = 0.03).
Figure 2.
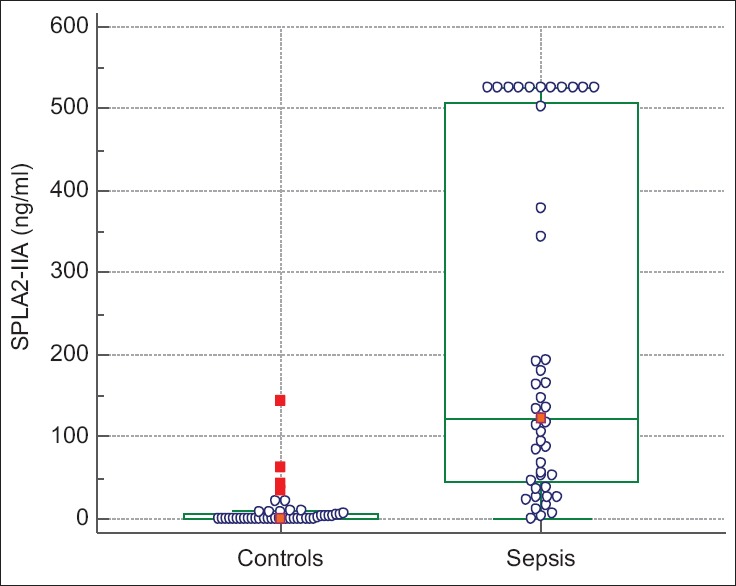
Box-and-whisker plot of median and interquartile ranges of secretory phospholipase A2-IIA levels in control and septic patients
Optimal cutoff threshold for the identification of sepsis using sPLA2-IIA appeared to be 25 ng/ml, with a sensitivity of 86.7% (confidence interval 72.5–94.5) and a specificity of 91.1% (confidence interval 77.9–97.1). Utilizing a higher cutoff of 100 ng/ml improved specificity to 97.8% (confidence interval 86.8–00.9) but with sensitivity of 57.8% (confidence interval 42.2–72.0). There were insufficient numbers to adequately determine optimal thresholds for determination of sepsis with a bacterial source versus patients meeting SIRS criteria with no bacterial source identified.
DISCUSSION
Optimal identification of septic patients in the ED is an important goal to decrease morbidity and mortality from this disease.[18] In this study, sPLA2-IIA was elevated significantly in patients presenting with sepsis to the ED as compared to controls with other medical illnesses. With an optimized cutoff of 25 ng/ml, this marker is sensitive and specific enough to be clinically useful in distinguishing the sepsis group from the control group in our ED population. Our findings are consistent with Tan et al., who noted that sPLA2-IIA was an excellent marker for identifying sepsis and accurately distinguished sepsis from nonsepsis groups (sensitivity 91%, specificity 78%, PPV 95, NPV 64%).[15] All patients in their study, however, met at least two SIRS criteria, and our data add to this body of literature by utilizing sPLA2-IIA assay in other ED patients who do not meet SIRS criteria.[15] At this point, it appears that 25 ng/mL may be an optimal cutoff with our assay which maximizes both specificity and sensitivity of this assay. In Tan's study, 2.3 μg/l (2.3 ng/mL) was the optimal cutoff.[15] This is likely due to the difference of capture and detection antibodies used in that particular assay. Previous studies that used other ELISA assays have shown ranges more similar to what we had found. In a study by Rintala et al., for example, they had found that sPLA2-IIA levels ranged from 1.32 to 25.25 ng/ml in normal patients and 12.95 to 1574 ng/ml in those presenting with sepsis.[16]
Rintala et al. found that sPLA2-IIA levels correlated with severity of disease and could indicate bacterial source of infection.[16] sPLA2-IIA levels may be indicative of disease severity and those with high initial levels may be strongly considered to be admitted to a higher level of care.[16] Our data support this with those presenting with a confirmed bacterial source of infection tending to have higher sPLA2-IIA levels than those with no identifiable bacterial source or a viral source of their illness. However, our pilot study was not designed to identify a clear relationship between sPLA2-IIA levels and severity of disease. With a larger patient sample population, our study may have demonstrated this same relationship with severity as Rintala. Nevertheless, elevated sPLA2-IIA levels may be useful in clinical practice to help identify a high-risk group of individuals with likely bacterial disease who may warrant admission to a higher level of care or inpatient admission rather than outpatient observation.
In our study, sPLA2-IIA levels had a weak relationship with ED lactate levels. Elevated lactate levels are used as a marker for severe sepsis and septic shock as per the Center for Medicare and Medicaid Services and the surviving sepsis campaign guidelines but may be elevated in other hypoperfused states in the absence of sepsis or may be elevated secondary to reduced lactate clearance.[19,20] Nevertheless, patients meeting sepsis criteria are resuscitated as per the specific guidelines based on initial lactate level. The weak relationship between initial ED lactate and initial ED sPLA2-IIA leaves many questions unanswered. Should septic patients with elevated sPLA2-IIA be resuscitated in a similar manner to septic patients with elevated lactic acid levels? Lactic acid is trended and clearance of lactate is used as an endpoint of resuscitation. Will trending sPLA2-IIA yield the same findings? Is sPLA2-IIA more specific than lactate in sepsis diagnosis? Despite our initial sPLA2-IIA tending toward higher levels with higher severity, the lack of a relationship with lactate in our data does not allow us to postulate on these unanswered questions. Certainly, further study is warranted.
We also noted that initial sPLA2-IIA had a weak relationship with length of stay. Length of stay as a surrogate marker of severity of disease is debatable, given the multifactorial nature of disposition decisions and timing. Further study utilizing clinical markers of disease severity, such as acute physiology and chronic health evaluation II or sequential organ failure assessment scoring for the sickest patients, would be helpful and likely shed some light on the relationship between sPLA2-IIA levels, disease severity, and disease progression.
Limitations
This study has several limitations. It is a small, pilot trial with a limited number of patients presenting to a single center. These patients were enrolled on a convenience basis, which may introduce bias. In addition, because our study required written consent before enrollment, some of the sickest patients presenting with sepsis were excluded because they were too ill to undergo consent. This explains our low mortality rate compared to national data. This sicker patient population, who would benefit most from early detection and prognostic information, requires further study.
Our specimen handling was a limitation, as well. There was a lag period between when samples were collected and when they were processed, as they had to be picked up and transported to an outside laboratory. Optimizing pick-up timing and sample storage (refrigerated versus room temperature, upright in test tube holders versus test tubes lying on their sides) resulted in hemolysis and exclusion of several samples. Therefore, the cutoff value for sPLA2-IIA may have better sensitivity and specificity than we have demonstrated if testing is performed under ideal conditions.
Finally, blood was drawn as part of a medical evaluation, and although all phlebotomy was done by trained medical staff, there could be subtle differences in technique that are not accounted for in this study.
CONCLUSION
Our study is in agreement with the previously published reports that show sPLA2-IIA to have potential as a marker in early diagnosis of patients presenting with possible sepsis. This marker appears to be sensitive and specific in distinguishing “sick” patients, especially those with a bacterial source of infection. This holds true in infections of multiple different organ systems. Further study is warranted to determine threshold value for severe sepsis, prognostic value, and role of trending sPLA2-IIA in the inpatient management of the septic patient.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–54. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR, et al. Epidemiology of severe sepsis in the United States: Analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29:1303–10. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Silva E, Pedro Mde A, Sogayar AC, Mohovic T, Silva CL, Janiszewski M, et al. Brazilian sepsis epidemiological study (BASES study) Crit Care. 2004;8:R251–60. doi: 10.1186/cc2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. [Last accessed on 2017 Mar 06]. Available from: https://www.cdc.gov/sepsis/datareports/index.html .
- 5.Rezende E, Silva JM, Jr, Isola AM, Campos EV, Amendola CP, Almeida SL. Epidemiology of severe sepsis in the emergency department and difficulties in the initial assistance. Clinics (Sao Paulo) 2008;63:457–64. doi: 10.1590/S1807-59322008000400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang HE, Shapiro NI, Angus DC, Yealy DM. National estimates of severe sepsis in United States emergency departments. Crit Care Med. 2007;35:1928–36. doi: 10.1097/01.CCM.0000277043.85378.C1. [DOI] [PubMed] [Google Scholar]
- 7.Rangel-Frausto MS, Pittet D, Costigan M, Hwang T, Davis CS, Wenzel RP, et al. The natural history of the systemic inflammatory response syndrome (SIRS). A prospective study. JAMA. 1995;273:117–23. [PubMed] [Google Scholar]
- 8.Clyne B, Olshaker JS. The C-reactive protein. J Emerg Med. 1999;17:1019–25. doi: 10.1016/s0736-4679(99)00135-3. [DOI] [PubMed] [Google Scholar]
- 9.Tang BM, Eslick GD, Craig JC, McLean AS. Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: Systematic review and meta-analysis. Lancet Infect Dis. 2007;7:210–7. doi: 10.1016/S1473-3099(07)70052-X. [DOI] [PubMed] [Google Scholar]
- 10.Thomas-Rueddel DO, Poidinger B, Weiss M, Bach F, Dey K, Häberle H, et al. Hyperlactatemia is an independent predictor of mortality and denotes distinct subtypes of severe sepsis and septic shock. J Crit Care. 2015;30:439e1–6. doi: 10.1016/j.jcrc.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 11.Borthwick HA, Brunt LK, Mitchem KL, Chaloner C. Does lactate measurement performed on admission predict clinical outcome on the Intensive Care Unit? A concise systematic review. Ann Clin Biochem. 2012;49:391–4. doi: 10.1258/acb.2011.011227. [DOI] [PubMed] [Google Scholar]
- 12.Lambeau G, Gelb MH. Biochemistry and physiology of mammalian secreted phospholipases A2. Annu Rev Biochem. 2008;77:495–520. doi: 10.1146/annurev.biochem.76.062405.154007. [DOI] [PubMed] [Google Scholar]
- 13.Hong CW, Kim TK, Ham HY, Nam JS, Kim YH, Zheng H, et al. Lysophosphatidylcholine increases neutrophil bactericidal activity by enhancement of azurophil granule-phagosome fusion via glycine. GlyR alpha 2/TRPM2/p38 MAPK signaling. J Immunol. 2010;184:4401–13. doi: 10.4049/jimmunol.0902814. [DOI] [PubMed] [Google Scholar]
- 14.Movert E, Wu Y, Lambeau G, Kahn F, Touqui L, Areschoug T, et al. Secreted group IIA phospholipase A2 protects humans against the group B streptococcus: Experimental and clinical evidence. J Infect Dis. 2013;208:2025–35. doi: 10.1093/infdis/jit359. [DOI] [PubMed] [Google Scholar]
- 15.Tan TL, Ahmad NS, Nasuruddin DN, Ithnin A, Tajul Arifin K, Zaini IZ, et al. CD64 and group II secretory phospholipase A2 (sPLA2-IIA) as biomarkers for distinguishing adult sepsis and bacterial infections in the emergency department. PLoS One. 2016;11:e0152065. doi: 10.1371/journal.pone.0152065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rintala EM, Nevalainen TJ. Group II phospholipase A2 in sera of febrile patients with microbiologically or clinically documented infections. Clin Infect Dis. 1993;17:864–70. doi: 10.1093/clinids/17.5.864. [DOI] [PubMed] [Google Scholar]
- 17.Mansour KM, Kuypers FA, Wang TM, Miller AM, Larkin SK, Morris CR. Secretory phospholipase A2: a marker of infection in febrile children presenting to a pediatric ED. Am J Emerg Med. 2011;29:1163–8. doi: 10.1016/j.ajem.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivers E, Nguyen B, Havstad S, Ressler J, Muzzin A, Knoblich B, et al. Early goal-directed therapy in the treatment of severe sepsis and septic shock. N Engl J Med. 2001;345:1368–77. doi: 10.1056/NEJMoa010307. [DOI] [PubMed] [Google Scholar]
- 19. [Last accessed on 2017 Mar 06]. Available from: https://www.cms.gov/Medicare/Quality-initiativespatient-assessment-instruments/qualitymeasures/core-measures. html .
- 20.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Crit Care Med. 2017;45:486–552. doi: 10.1097/CCM.0000000000002255. [DOI] [PubMed] [Google Scholar]


