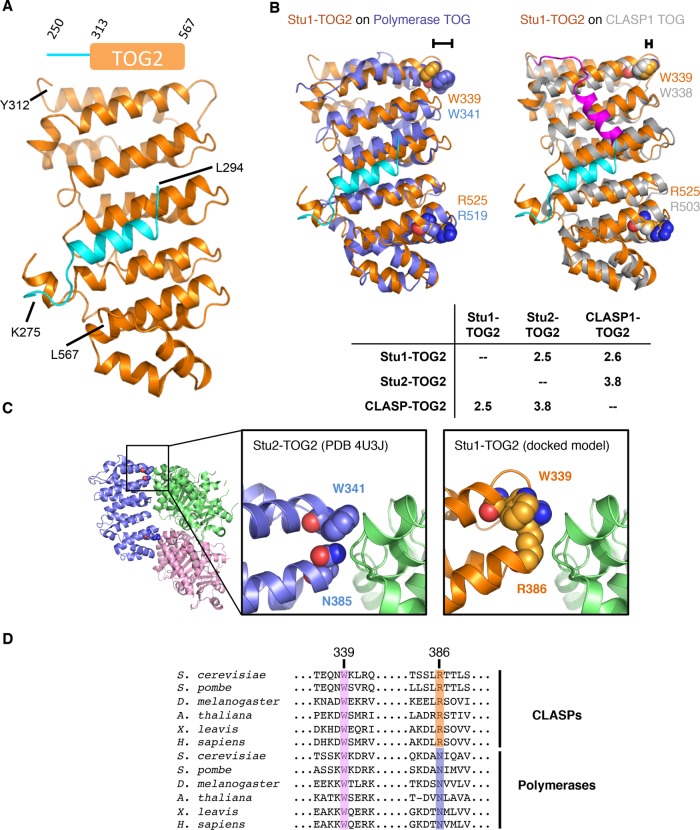
FIGURE 3:
Crystal structure of Stu1-TOG2. (A) Cartoon representation of the TOG2 structure (orange; the linker sequences are in cyan). Inset shows the construct crystallized (repeated from Figure 1). (B) Rigid body superpositions of Stu1-TOG1 (orange; docked linker, cyan) onto a polymerase TOG (left, Stu2-TOG2, slate) or a CLASP TOG (right, CLASP1-TOG2, gray; docked linker, magenta). Conserved Arg and Trp residues implicated in tubulin binding are shown as spheres and colored to match their respective TOG. The different length brackets illustrate the difference in positioning of the conserved tryptophan. Cα rms coordinate deviation values for the superpositions are presented in the inset table. (C) A distinctive tubulin-binding interface for Stu1-TOG2. Cartoon representation of the Stu2-TOG2:tubulin complex (left; PDB 4U3J), with a region of interest boxed. Close-up view of the TOG:tubulin interface in the region of interest for Stu2 (middle) and Stu1 (right). In Stu1-TOG2 an Arg fills the space normally taken by a tryptophan; this Arg likely contacts tubulin directly, conferring distinctive tubulin-binding properties. (D) Multiple-sequence alignment of CLASP-family (top) and polymerase-family (bottom) TOG domains. W339 (Stu1 numbering) is highly conserved in both CLASP and polymerase TOGs; R386 is highly conserved in CLASPs but in polymerases that position is an asparagine.