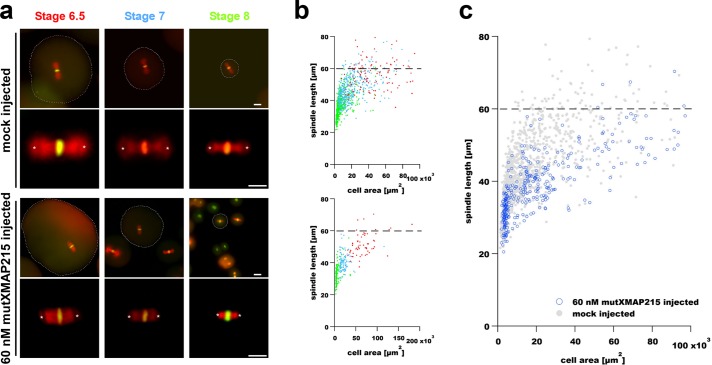
FIGURE 2:
Microinjection of a polymerase-deficient mutant XMAP215 decreases spindle length in early X.laevis embryos. (a) Images show representative spindles in blastomeres isolated from embryos at the indicated stages. The two top rows show spindles in blastomeres from control (mock-injected) embryos, whereas the two bottom rows show spindles from blastomeres isolated from embryos injected with 60 nM mutXMAP215. Spindle MTs and DNA were labeled as in Figure 1. Scale bar = 50 µm (images in first and third rows). Scale bar = 30 µm (higher-magnification spindle images in second and fourth rows). Spindle length was again measured as the aster center–to–aster center distance (centers are marked with white asterisks). Cell boundaries, indicated by dashed outlines shown in lower-magnification spindle images, were manually traced and used to determine cell cross-sectional area for scaling plots. (b) Spindle length measurements are shown plotted as a function of blastomere cross-sectional area in mock-injected embryos (top plot) and in embryos injected with 60 nM mutXMAP215 (bottom plot). Data point colors correspond to stage-specific text colors shown above images in panel a. (c) Data from both graphs in b were plotted in a single graph and show a general downward shift in spindle length with the injection of 60 nM mutXMAP215. Dashed horizontal lines indicate the putative upper limit to spindle size in X. laevis embryos (Wuhr et al., 2008).