Figure 2. ZNFX-1 helicase and cysteine-rich domains are required for epigenetic inheritance.
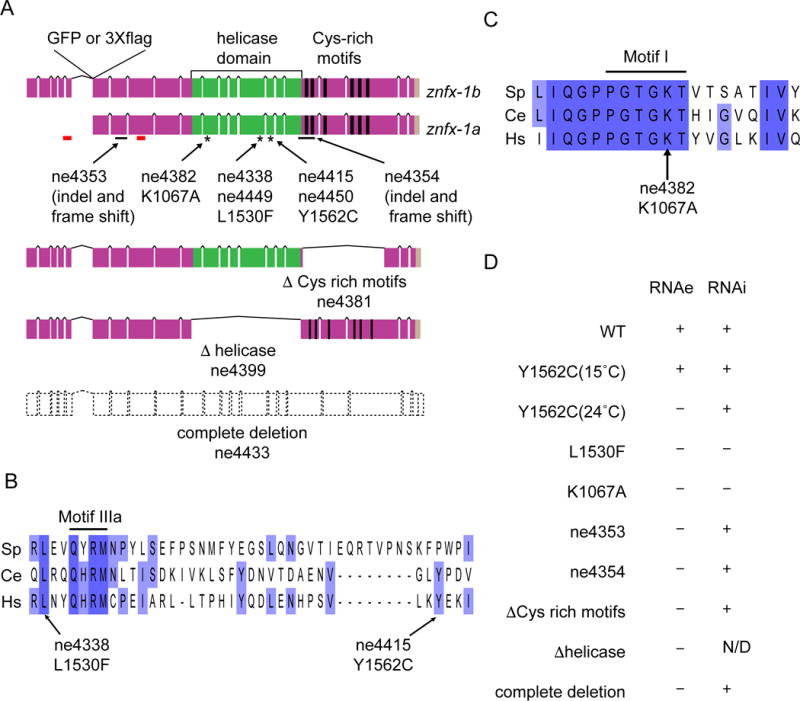
(A) Schematic diagram depicting the structure of znfx-1 locus and locations of predicted domains. Positions of GFP or 3×flag tag insertion, indel (solid black lines) or point (asterisks) mutations used in the study, regions targeted in RNAi in Figure S3B (red lines), as well as the structures of in-frame mutations deleting Cys-rich motifs or helicase domain and of complete deletion mutation, are shown.
(B) Alignment of a region encompassing conserved motif IIIa of helicase domains (solid black line) from Schizosaccharomyces pombe (Sp), Caenorhabditis elegans (Ce) and Homo sapiens (Hs), and the positions of point mutations identified in the screen are shown. Purple, lighter purple, and no background indicate 100%, 75%, 0% conservation.
(C) Alignment of a region encompassing conserved motif I (solid black line) of helicase domains represented in (B). The position of K1067A mutation used in the study is shown.
(D) Table summarizing the phenotypes of alleles used in the study. Defects in RNAe or cold sensitive RNAi are indicated by + (wild-type) or - (defective). N/D represents (not determined). See also Figures S2 and S3.
