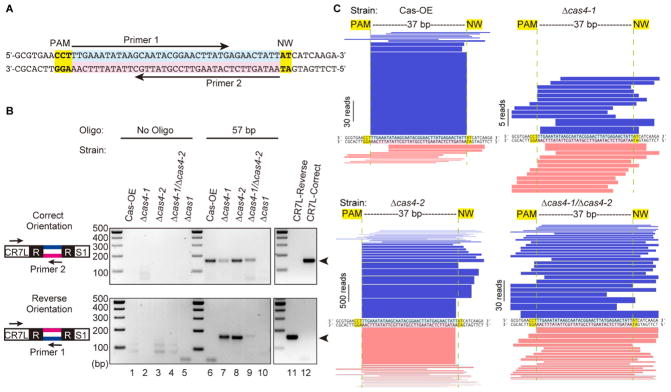
Figure 3. Cas4-1 and Cas4-2 trim and orient spacers.
(A) Diagram of the 57 bp duplexed DNA oligonucleotide used as a protospacer in adaptation assays. Arrows indicate the locations of primers used in PCR for detecting oligo integration into the array. (B) Analysis of oligo-specific integration. PCR products are amplified using primers binding to CRISPR7 leader and oligo-specific primers in either the forward or reverse orientation. Sizes of DNA standards are indicated. The PCR product corresponding to leader-integrated oligo is indicated with an arrowhead. (C) Visual representation of integrated 57 bp oligos. The DNA sequence in the middle shows the 57 bp oligo as in part a. Blue bars show the fragments of the oligo that were found integrated into the array in the correct orientation with respect to the PAM while pink bars show the fragments that were integrated in the reverse orientation. Representations correspond to data that were pooled from 16 experiments (CRISPR5 and CRISPR7 arrays, 8 replicates each, see Table S2 for raw values).