Abstract
Electronic health record (EHR) data are becoming a primary resource for clinical research. Compared to traditional research data, such as those from clinical trials and epidemiologic cohorts, EHR data have a number of appealing characteristics. However, because they do not have mechanisms set in place to ensure that the appropriate data are collected, they also pose a number of analytic challenges. In this paper, we illustrate that how a patient interacts with a health system influences which data are recorded in the EHR. These interactions are typically informative, potentially resulting in bias. We term the overall set of induced biases informed presence. To illustrate this, we use examples from EHR based analyses. Specifically, we show that: 1) Where a patient receives services within a health facility can induce selection bias; 2) Which health system a patient chooses for an encounter can result in information bias; and 3) Referral encounters can create an admixture bias. While often times addressing these biases can be straightforward, it is important to understand how they are induced in any EHR based analysis.
Introduction
The gold standard data source for clinical research are clinical trials and epidemiological cohort studies [1]. Clinical trials are governed by protocols and are carefully organized to answer discrete questions. A well-designed trial systematically collects data from participants ensuring the same information is collected for all participants. It is clear who is in the trial, when the intervention begins, and how the data are collected, resulting in appropriate information capture. Similarly epidemiological cohorts draw from a well-defined population, with meaningful time windows and pre-specified data fields, though participants are not randomized to interventions as in trials. However, clinical trials and epidemiological cohorts are time and cost intensive. For these reasons, there has been significant work recently using electronic health record (EHR) data for clinical research.
Data from the EHR are an attractive research tool because of the volume and velocity of data emanating from it, the quintessential clinical ‘big data.’ As such, EHRs allow researchers to flexibly study multiple clinical outcomes. However, since they are designed for clinical care and billing purposes, rather than research, the data from them are inherently different from traditional data sources [2]. They present an opportunistic and non-random snapshot of patient interactions, capturing only the information that is relevant to each specific encounter and across a variety of contexts that are represented by how a patient interacts with the health system.
A patient may interact with a health system for a number of reasons across different settings, generating new data with each interaction. These variable interactions can have downstream effects biasing data analyses. We have previously referred to these potential biases as ‘informed-presence,’ the idea that people do not interact randomly with a health system [3]. In this paper, we aim to illustrate the various ways that a patient’s interaction with the health system can impact standard analyses. We illustrate this via three vignettes that utilize real world EHR data drawn from our collaborative work. In doing this, we demonstrate how selection into the EHR can impact seemingly standard analytic questions through examples of selection bias, information bias, and admixture bias. Our underlying premise is that many of the biases associated with EHR data can be mitigated via appropriate and thoughtful study design.
EHRs as a Missing Data Problem
Over the past few years a considerable amount of clinical research has utilized EHR data [4,5]. Though EHRs are a valuable research tool, [6,7] analytical challenges dealing with confounding and selection bias have been observed [8]. Much of this research has been framed as a problem of missing data [9] which stems from the presence of unstructured data and a general assumption that missing values equate to negative results.
The impact of this incomplete data has been studied in various contexts: diagnoses, laboratory values and vitals measurements. Rates of missingness vary across populations, potentially biasing estimates based on available data and even entire analytic samples [10,11]. Approaches to deal with these challenges have been proposed but have not been adopted uniformly across analyses [4]. In many cases, missing EHR data cannot be ignored or simply imputed based on the assumption that data are missing at random [12].
The missing data problem is further complicated when considering the multiple mechanisms responsible for creating EHR data (e.g. patient interactions, workflow). Since the health record captures the clinical workflow in addition to the patient’s pathology, one must consider both elements when analyzing EHR data [13]. The mechanisms responsible for creating EHR data, along with the sparse nature of EHR data, can make standard methods of dealing with missing data invalid or inappropriate [8,14]. Leveraging the contextual information of an encounter (i.e., reason for lab measurement or if the encounter was patient initiated) can improve analysis [14,15].
Thinking about the causes and consequences of bias due to missing data is standard practice within epidemiology. Since patients do not interact with a health system randomly we prefer to place the emphasis not on what is missing, but what is observed. In previous work, we illustrated that the number of interactions a patient has with a health system, can induce either Berkson’s bias or M-Bias [3]. In this paper, we extend this framework to other characteristics of patient interactions within a health system (Figure 1). This is important as informed presence corresponds to a range of biases with respect to analyzing EHR data. Overall, the following vignettes illustrate that where interactions occur inform the data collected and therefore impact the corresponding analysis. While the solution can be straightforward – often stratification is effective – ignoring these effects can result in biased associations.
Figure 1.
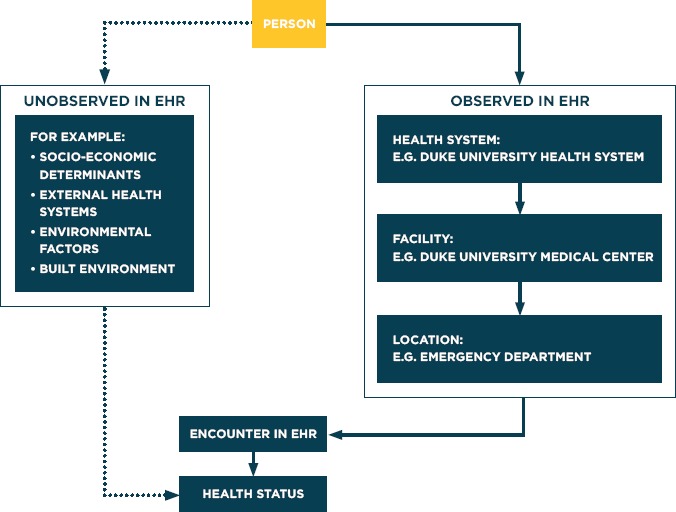
The Flow of Patient Interactions to Create EHR Data Representative of Health Status
Data Used
The data for this study are derived from the Duke University Health System (DUHS). DUHS consists of three hospitals – a large referral hospital and two community hospitals – as well as a network of outpatient clinics. It is estimated that 85 percent of Durham County residents receive health care services at DUHS [16]. The DUHS EHR system is an EPIC based system installed incrementally from 2012 through 2013, with pre-EPIC data going back to 1996.
For the primary analyses, we used a datamart that includes EHR data from 2007-2014 which follows the PCORNet v3.0 Common Data Model [17]. This datamart contains encounter specific information on patient demographics (e.g. age, sex, race), vital measurements (e.g. systolic and diastolic blood pressure, weight), laboratory values (e.g., hemoglobin A1C (HbA1c), glucose) and medications. In total, 267,375 unique patients were available for analysis, covering 7,387,526 encounters.
For each of our examples we supplemented our primary EHR database with data from three additional data sources. First, we linked data from a community intervention trial of high risk diabetic patients [18] with biometric measurements taken during home health visits by community health workers. Second, we linked DUHS patient data to the EHR of Lincoln Community Health Clinic (LCHC). LCHC is a federally qualified health clinic providing primary care to under-served populations within Durham County Finally, we linked data from a database containing information on patients who received a catheterization at the Duke Heart Catheterization Laboratory from 2010 to the present [19,20].
Illustration of Potential Biases
Location Within Facility: Selection Bias
The data collected in trials have mechanisms in place designed to minimize biases [21]. These mechanisms include scheduled encounters and pre-specified protocols that define the information that is to be collected at each encounter. However real-world medical encounters occur informatively with respect to an individual’s health. Moreover, patients can engage with the health system across a variety of settings, including outpatient visits, inpatient stays, and emergency department (ED) visits. Encounters within these settings are all captured within a single EHR system. Each of these encounters may collect the same type of information (e.g. blood pressure, laboratory measurements etc.); however, the context of the encounters will vary greatly leading to informed presence and Berkson’s bias.
To illustrate this, we consider blood pressure, which is frequently collected across different locations within a health system. We performed this analysis among 171 diabetic patients in a community health intervention trial [22]. For each patient EHR data were extracted from the emergency department (ED), inpatient, and outpatient settings. Additionally data were collected during quarterly home visits, providing multiple measurements per-person. For our purposes, we consider the home visits to be analogous to clinical trial encounters since they were prescheduled and therefore non-informative with respect to the individual’s health. Using a random effects model to account for within person correlation, we estimated mean systolic blood pressure across the four locations (Figure 2). We found that blood pressure measured in the ED is clinically and statistically higher than at home (145.4 mmHg vs 137.3 mmHg, respectively P < 0.001). Additionally the ED measurements show greater variability as compared to home visits, inpatient encounters, and outpatient encounters (standard deviation = 25.8, 21.1, 21.7, and 23.9, respectively; P < 0.001). Conversely, the outpatient values were comparable to the values from home visits suggesting that they may not be informative and potentially more appropriate for use in analyses. While it is common knowledge among clinicians that patients have higher blood pressure measurements in the ED, [23] these differences need to be accounted for in any analysis.
Figure 2.
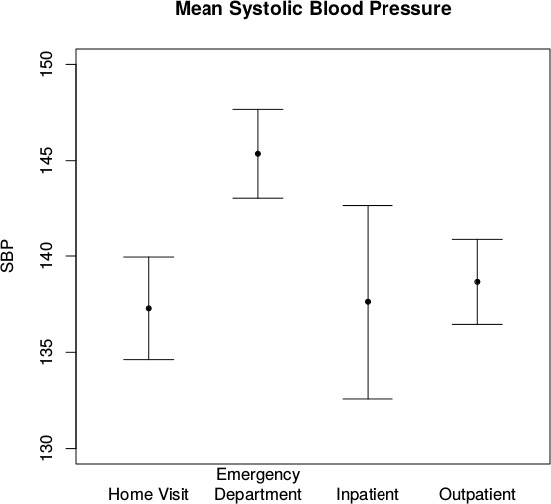
Collection of Mean Systolic Blood Pressure in Diabetic Patients Across Health System Facilities
We used a larger population of diabetic patients to illustrate the impact of not accounting for these differences. We assessed the association between HbA1c values and risk of myocardial infarction (MI), identifying a sample of 11,873 diabetic patients within the Duke EHR from 2007–2011. Among these patients, we identified the time to first MI from 2012–2014, administratively censoring at 12/31/2014. During this period, 3,789 HbA1c measurements were taken in the ED setting, and 46,743 were taken in the outpatient setting. Median HbA1c values across locations differed significantly: 7.7 percent and 7.0 percent for ED and outpatient observations, respectively (P < 0.001). Using a time-varying Cox model, we found that higher HbA1c levels are associated with increased risk of MI (hazard ratio (HR) =1.13, 95 percent confidence internal (CI): 1.07, 1.19) when location is not included in the model, confirming the known relationship between HbA1c and myocardial infarctions (Table 1 and 2) [24]. Adjusting for location attenuates the association (HR= 1.02: 95 percent CI: 0.97, 1.08), suggesting that location may confound the association between HbA1c and MI. However, testing for an interaction between HbA1c and location resulted in a significant term, indicating that there is a differential effect based on location.
Table 1.
Hazard Ratio for 1% Increase in Hemoglobin A1c Stratified by Where HbA1c was Measured
| HAZARD RATIO | 95% Cl | P-VALUE | |
|---|---|---|---|
| All Labs – Unadjusted for Location | 1.13 | 1.07, 1.19 | <0.001 |
| All Labs – Adjusted for Location | 1.02 | 0.97, 1.08 | 0.37 |
| Outpatient Labs Only | 1.13 | 1.06, 1.21 | <0.001 |
| Emergency Department Labs Only | 0.96 | 0.90, 1.02 | 0.16 |
Abbreviations: CI, Confidence Interval.
aAdjusted for age, sex, race.
Table 2.
Odds Ratios of Cancer Diagnosis Based on Diabetes Status, Startfied by Locationa
| LOCATION | ODDS RATIOa | 95% CI |
|---|---|---|
| All | 1.69 | 1.36, 2.10 |
| DUMC-Only | 1.46 | 1.15, 1.87 |
| DRH-Only | 0.89 | 0.63, 1.26 |
| LCHC-Only | 1.08 | 0.74, 1.56 |
Abbreviations: CI, Confidence Interval; DUMC, Duke University Medical Center; DRH, Duke Regional Hospital; LCHC, Lincoln Community Health Center.
aAdjusted for age, sex, race, and the number of encounters.
To further assess the impact of location, we re-ran the analysis stratifying HbA1c measurements based on whether they were taken in the outpatient or ED settings. The outpatient results are similar to our overall assessment with a HR = 1.13, 95 percent CI: 1.06, 1.21, though the HbA1c effect is not apparent in the ED (HR=0.96, 95 percent CI: 0.90, 1.02). We hypothesize that there is a selection effect bringing people into the ED, biasing the effect towards the null. That is, patients who have their HbA1c measured in the ED may have differential risk for MI as compared to patients who have their HbA1c measured in an outpatient setting. In our example, patients with unmanaged diabetes (high HbA1c) may be more likely to have a non-urgent ED visit as compared to patients with managed diabetes who visit the ED [25]. Therefore, where someone seeks care can be informative of their health status.
Facility Within Health System: Information Bias
One useful feature of trials and epidemiological cohorts is that all patients have the same information collected. As mentioned before, data within the EHR are only collected if they are considered clinically important at the time of the encounter [8]. For this reason, patients with diabetes have more recorded HbA1c measurements within their EHR than patients without diabetes [10]. This may not necessarily bias the results if the analytic sample includes only diabetic patients. However, information bias can be present if we only use data from a subset of the facilities utilized by a patient who seeks care across multiple facilities. This was shown within a VA population; patients who use both VA and non-VA services had fewer measured comorbidities when only a single health system EHR was used in analyses [26]. This can impact analyses of an EHR based study in multiple ways, including defining a cohort, phenotyping comorbid conditions, and recording outcomes. In trials or observational studies, these components are well defined a priori with any data element needed to define an outcome or a comorbidity collected. However, for EHR based analyses outcomes must be derived using the available data, and the choice of data may ultimately impact the final ‘phenotype.’ [27,28] In earlier work we showed that differential quality of clinical phenotyping, via number of patient encounters, can bias associations [3]. We further explore this with respect to seeking care at multiple facilities.
In this example, we consider patients seeking care across the Duke Health System, including the Duke University Medical Center (DUMC), Duke Regional Hospital (DRH) and Lincoln Community Health Clinics (LCHC). LCHC contains exclusively outpatient encounters whereas DUMC and DRH include inpatient, outpatient, and ED encounters.
We identified 9,568 patients that had encounters within all 3 facilities from 2012 and 2013. To demonstrate differences in the facilities’ data capture, we identified patients with two chronic diseases: diabetes using ICD-9 codes in conjunction with medications and laboratory values for HbA1c or Glucose (Random or Fasting), [28] and any cancer using ICD-9 codes [29].
Figure 3 shows the disease classifications for the 9,568 Durham County residents that received care at all three locations. If the same information was collected across all facilities, then all of the disease positive cases would be at the intersection of the three facilities in the Venn diagrams. This is not the case for cancer where only 9.9 percent are captured at all facilities. This implies that if you were to classify a patient’s cancer status using only the data from one facility you would have a high probability of misclassifying them. One reason for diseases to differ across facilities is that patients are going to each facility for different reasons. It is not surprising that most of the cancer diagnoses occur at DUMC (83 percent) which has a nationally recognized cancer center compared to DRH (37 percent) or LCHC (33 percent) (n = 477). However, when we look at a condition such as diabetes which can be managed by different specialists and across multiple facilities, we see rates that are more comparable across facilities—DUMC: 73 percent; DRH: 59 percent; LCHC: 72 percent (n= 2,783).
Figure 3.
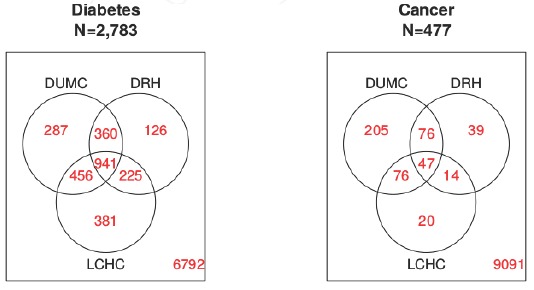
Venn Diagram of Phenotype Classification When Restricting Available Data to Individual Faculties
Abbreviations: DUMC, Duke University Medical Center; DRH, Duke Regional Hospital; LCHC, Lincoln Community Health Center.
We explored this further by considering an analysis assessing the association between the two disease states. Using the phenotypes above we ran a logistic regression model to quantify the odds of having diabetes based on cancer status. For illustrative purposes, we minimally adjusted for age, sex, race, and the number of encounters [3] during the study. When we use data from all three facilities we see that the odds for diabetes are 69 percent higher when a patient has cancer (Odds ratio=1.69, 95 percent CI: 1.36, 2.10). However, when the location-specific phenotypes are used in our logistic regression the odds ratios are 1.46, 0.89, and 1.08 for DUH, DRH, and LCHC respectively. On the same population, we get very different results for the associations based exclusively on where the data are coming from. Since a minority of cancer patients receive cancer-related care at DRH or LCHC, when conducting analyses related to cancer, only using data from either facility may produce biased results.
External Health Systems and Referral Patients: Admixture Bias
A final consideration is being able to define the study’s source population. An important characteristic of clinical trials and observational studies is that all patients come from the same clinical population-even if it may be poorly defined. Conversely, patients seen by a medical system, may constitute a well-defined source population-people seeking care at DUMC-but can constitute a mixture of clinical populations. Although admixtures have a specific context in genetics, [30] we see a natural extension to the medical system population. This becomes an important challenge for systems like DUMC that serve both a local and a referral population. The referral population is often meaningfully different from the local population.
Referral patients may be sicker or have a different distribution of underlying comorbidities. This is exacerbated by the fact that referral patients often only contribute inpatient data because they receive outpatient care through an institution closer to their home. This can lead to the two previously discussed biases: selection bias and information bias. However, a third bias (i.e., admixture bias) emerges as well; by the sheer nature of being referred, the patient population is different.
We investigated this through a database maintained by the Duke Heart Catheterization Laboratory, which sees patients who are referred and those who receive the majority of their care through the Duke Health System. Since the catheterization laboratory collects routine information on all patients, the potential for the previously discussed biases is minimized. We used patient address to determine whether a patient was a referral patient or not, defining referral patients as those residing > 50 miles from DUMC and non-referral patients as residents of Durham County, from 2010-2014. During this period 2,114 patients were non-referral patients and 5,522 were referral. Examining comorbidities and catheterization results stratified by referral status, we observed two distinct populations (Table 3). Non-referral patients were more likely to be Black (49.6 percent vs 24.1 percent), have a higher history of MI (23.4 percent vs 15.6 percent), hypertension (68.5 percent vs 57.6 percent), and angina (63.6 percent vs 43.8 percent). Based on the catheterization results more referral patients have a confirmation of valvular heart disease (7.0 percent vs 3.8 percent), and moderate-to-severe aortic (20.0 percent vs 7.3 percent) and mitral valve stenosis (61.9 percent vs 48.0 percent) among patients that had their level of stenosis measured. Conversely non-referral patients are more likely to have a history of progressive vascular disease. Clinically, this is not surprising. Patients with atypical or more severe valvular diseases tend to merit a referral to a specialty center such as the Duke Heart Catheterization Laboratory, whereas the less complex vascular cases are less likely to warrant a referral. Differences among outcomes confirm that these are two distinct populations. The referral patients had higher rates of mortality (HR: 1.52; 95 percent CI: 1.34, 1.72) (Fig 4) and valve repair (HR: 1.82, 95 percent CI: 1.49, 2.21) and lower rates of Percutaneous Coronary Intervention (HR: 0.61; 95 percent CI: 0.55, 0.67) compared to the non-referred population.
Table 3.
Baseline Demographics and Clinical Characteristics of Catheterization Patients by Referral Status
| CHARACTERISTIC | REFERRAL PATIENTS (N = 5522) | NON-REFERRAL PATIENTS (N = 2114) | P-VALUE |
|---|---|---|---|
| Age (years)(Median, 25th–75th Quartile) | 64 (54–71) | 61 (52–71) | 0.16 |
| Female | 2317/5522 (42.0%) | 947/2114 (44.8%) | 0.03 |
| White | 4138/5449 (75.9%) | 1054/2093 (50.4%) | <0.001 |
| COMORBIDITIES | |||
| Myocardial Infarction | 859/5522 (15.6%) | 495/2114 (23.4%) | <0.001 |
| Renal Disease | 336/5522 (6.1%) | 158/2114 (7.5%) | 0.03 |
| Hypertension | 3180/5522 (57.6%) | 1448/2114 (68.5%) | <0.001 |
| Hyperlipidemia | 2414/5522 (43.7%) | 1001/2114 (47.4%) | 0.004 |
| Angina | 2412/5512 (43.8%) | 1342/2109 (63.6%) | <0.001 |
| Congestive Heart Failure | 2253/5465 (41.2%) | 577/2092 (27.6%) | <0.001 |
| On Dialysis | 137/5522 (2.5%) | 82/2114 (3.9%) | 0.001 |
| VITALS | |||
| SBP mmHg (Median, 25th–75th) | 132 (118–147) | 139 (124–155) | <0.001 |
| CATHETERIZATION RESULTS | |||
| Valvular Heart Disease | 385/5522 (7.0%) | 80/2114 (3.8%) | <0.001 |
| 1 to 3 Diseased Vessels | 2084/3980 (52.4%) | 1031/1844 (55.9%) | 0.01 |
| Mitral Regurgitation Grade (I–IV) | 162/721 (22.5%) | 68/468 (14.5%) | <0.001 |
| Aortic Valve Insufficiency (Mild to Severe) | 155/298 (52.0%) | 35/88 (39.8%) | 0.04 |
| Aortic Valve Stenosis (Mild to Severe) | 185/719 (25.7%) | 38/342 (11.1%) | <0.001 |
| Left Ventricle Ejection Fraction(%) (Median, 25th–75th) | 59.00 (50.0-66.6) | 58.93 (49.8-66.0) | 0.98 |
| Interventional Coronary Cath | 731/5522 (13.2%) | 492/2114 (23.3%) | <0.001 |
| Diagnostic Coronary Cath | 4036/5522 (73.1%) | 1875/2114 (88.7%) | <0.001 |
| OUTCOMES | |||
| 5 Year Mortality | 1036/5522 (18.8%) | 328/2114 (15.5%) | <0.001 |
| Valve Surgery or Repair at 6 months | 537/5522 (9.7%) | 122/2114 (5.8%) | <0.001 |
| Percutaneous Intervention at 6 months | 899/5522 (16.3%) | 565/2114 (26.7%) | <0.001 |
Abbreviations: SBP, Systolic Blood Pressure.
Figure 4.
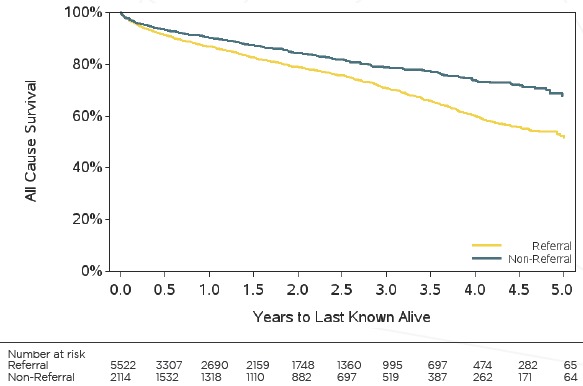
Kaplan Meier Curve of All-Cause Survival for Duke Catheterization Patients Stratified by Referral Status
While it is clear that there are two admixed populations, it is less clear how this may be addressed. The analytical perspective may inform how to proceed with these analyses. For example, from the health system perspective, it may be important to know survival statistics for patients at DUHS as a whole. In this scenario, combining the referred and non-referred populations seems appropriate. Conversely, from the patient perspective, such an analysis may not be appropriate, as it would result in a biased association, negatively impacting patient care. In this scenario, it may be helpful to stratify results by referral status.
Discussion
Our vignettes demonstrate that due to the informative nature of EHR data, care must be taken when using it for clinical research. The way that a patient interacts with a health system provides useful information to researchers on what data are present and why. The types of patient interactions may vary depending on the health system that is being used for research. Moreover, characteristics tied to these patient interactions may create biases that differentiate EHR data from clinical data sources like clinical trials or epidemiological cohorts. Understanding what data are available and contextual information linked to that data allows researchers to address biases appropriately through stratification.
When analyzing data from comprehensive medical centers, selection bias is the most likely bias to be encountered because these centers offer more than one type of patient service. While the clinical context of a laboratory measurement (i.e., diagnosing, screening, follow-up) is informative, [15] we see that the location of a laboratory or vital measurement can also have an impact on its value and the corresponding analysis, and is often easier to capture in the EHR. Disparate encounter types may record the same measurements but the context of the encounter may be informative. This selection bias is unique to EHRs as trials and observational cohorts control how data are received, reducing the confounding effect. In our example labs and vitals in the ED had both a different range of values and showed more variability than in other locations; outpatient measurements were comparable to measurements from non-random home visits. There are reasons why we may not want to use the ED data as an ED encounter may be inherently different than a scheduled outpatient encounter When we modeled the association of HbA1c with acute myocardial infarctions we saw different results when we used ED and outpatient data separately. An ED encounter may be informative of an additional signal impacting the data, for example the patient’s health status.
Moreover, in a region containing multiple health systems there is a possibility for information bias to impact analyses. A patient makes the decision of when and where to receive care, so they may interact with different health systems for different reasons. Understanding why patients have encounters in a given health system allows a researcher to confirm that the appropriate data is captured or potentially obtain additional data. Veterans tend to use Medicare rather than VA facilities to treat more serious illnesses [26]. Similarly we found patterns in utilization can explain why the same information appears at different rates across facilities, as seen with cancer diagnosis codes in our second vignette. The majority of cancer related encounters in Durham County occurred at the hospital with a cancer center. Without having access to the cancer center data, analyses would be biased. Knowing that there is a lack of information, the bias could be lessened by linking to an external data source [9].
Finally we described a previously unrecognized complication with EHR data, the potential for admixture. We observed that within the Duke Heart Catheterization Laboratory there are two distinct patient populations: referral and local. Referral patients are receiving catheterizations related to more complex valvular disease (higher levels of stenosis/regurgitation), consequently their outcomes and treatment is different than the non-referral patients. Referral patients also have higher rates of valve surgery and repair and higher rates of mortality. Since the two populations have different sets of risk factors, adjustment is necessary. An interesting complication, is that depending on the perspective of the analysis-health system or patient perspective-such admixture may or may not present a source of bias. Such scenarios are worthy of further consideration.
Much of the prior literature on biases associated with EHR data has focused on missing data problems. Our focus on what data are available and the associated biases should be viewed as complementary to previous work and as an aide to illustrate the complexity of EHR data. Analytic strategies to correct EHR data related biases are sensitive to what data are being analyzed [4]. By incorporating information related to how a patient interacts with the health system one can better understand the data. Appreciating the data quality issues and adjusting appropriately can transform the EHR into a suitable clinical research tool [31]. In order to effectively use an EHR for clinical research additional considerations must be made compared to a trials database.
This work is not meant to be exhaustive of challenges in designing EHR based studies, but present different considerations for EHR based analyses (see Box 1). Beyond the described patient interactions with a health care system, additional considerations arise. Extensive work has been done assessing how best to define patient outcomes and exposures, referred to as phenotyping [28,32]. Unlike medical claims data, EHR data allow one to consider multiple types of data elements for phenotyping as compared to claims data which mainly contains codes needed for billing. This allows the creation of more sensitive or specific definitions based on the needs of the researcher Another challenge is how best to define person-time [33]. A dataset created from EHR data constitutes an arbitrary cross-section of patient interaction with a health care system. Defining the baseline time point (i.e., time zero [t0]) or the time of disease incidence can be challenging. Often ‘burn-in’ periods (i.e., time prior to t0 that is used to define the study cohort) are required. Similarly, properly defining censoring and loss to follow-up may require ‘burn-out’ periods (i.e. time after an administrative censoring date).
There are important caveats to our analyses. Most importantly, our vignettes were designed to highlight challenges associated with analyzing EHR data. Therefore, the reported associations should not be construed as inference. Moreover, our examination of these issues is somewhat subjective and while these examples come directly from collaborative projects, their inclusion is prone to their own selection bias and are limited in scope to a single geographic region. Future work should aim to more systematically evaluate the existence and pervasiveness of these problems beyond our health system. Finally, our findings capture patient interactions in a static sense. A patient’s interactions and how they flow through a health system may evolve over time [34]. Understanding and capturing this process could provide additional information.
Box 1. Design Considerations for EHR Based Studies
Where in the health system are the data collected?
What is the coverage/catchment area of your health system?
Is the patient population receiving care across multiple institutions/centers?
Do the data constitute different catchments? (Admixture)
How are you defining exposures and outcomes? (Phenotyping)
-
How are you defining person-time?
What is an appropriate ‘burn-in’ period to define a cohort?
Is a ‘burn-out’ period necessary to define censoring?
Do different populations produce more information (i.e. sicker patients have more encounters)?
Considering the ways in which a patient interacts with a health system can be thought of as internal validation of the research question, ensuring that the question is being applied to the correct data. The ways in which a patient interacts with a health system can be informative of how EHR data should be used and understanding the context of an encounter helps a researcher decide the best analytic approach to take.
Acknowledgments
We thank the anonymous reviewer for his/her helpful comments. Research reported in this publication was supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) career development award K25 DK097279 (BAG) and National Institute of Diabetes, Digestive and Kidney Diseases of the National Institutes of Health (NIH) under Award Number P30DK096493 (NAB). Data extraction was supported by the National Center for Advancing Translational Sciences (NCATS), NIH, through Grant Award Number UL1TR001117 at Duke University. Data from SEDI was supported in part by Grant Number 1C1CMS331018-01-00 the Department of Health and Human Services, Centers for Medicare & Medicaid Services, and in part by the Bristol Myers Squibb Foundation Together on Diabetes program. The contents of this publication are solely the responsibility of the authors and have not been approved by the Department of Health and Human Services or the Centers for Medicare & Medicaid Services.
References
- 1.Bothwell LE, Greene JA, Podolsky SH, Jones DS. Assessing the Gold Standard--Lessons from the History of RCTs. N Engl J Med. 2016;374(22):2175–81. [DOI] [PubMed] [Google Scholar]
- 2.Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PR, Bernstam EV, et al. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013;51(8 Suppl 3):S30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein BA, Bhavsar NA, Phelan M, Pencina MJ. Controlling for Informed Presence Bias Due to the Number of Health Encounters in an Electronic Health Record. Am J Epidemiol. 2016;184(11):847–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dean BB, Lam J, Natoli JL, Butler Q, Aguilar D, Nordyke RJ. Review: use of electronic medical records for health outcomes research: a literature review. Med Care Res Rev. 2009;66(6):611–38. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein BA, Navar AM, Pencina MJ, Ioannidis JP. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc. 2017;24(1):198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Green BB, Anderson ML, Cook AJ, Catz S, Fishman PA, McClure JB, et al. Using body mass index data in the electronic health record to calculate cardiovascular risk. Am J Prev Med. 2012;42(4):342–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sagreiya H, Altman RB. The utility of general purpose versus specialty clinical databases for research: warfarin dose estimation from extracted clinical variables. J Biomed Inform. 2010;43(5):747–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haneuse S, Daniels M. A General Framework for Considering Selection Bias in EHR-Based Studies: What Data Are Observed and Why? EGEMS; (Wash DC: ). 2016;4(1):1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wells BJ, Chagin KM, Nowacki AS, Kattan MW. Strategies for handling missing data in electronic health record derived data EGEMS; (Wash DC: ). 2013;1(3):1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rea S, Bailey KR, Pathak J, Haug PJ. Bias in recording of body mass index data in the electronic health record. AMIA Jt Summits Transl Sci Proc. 2013;2013:214–8. [PMC free article] [PubMed] [Google Scholar]
- 11.Rusanov A, Weiskopf NG, Wang S, Weng C. Hidden in plain sight: bias towards sick patients when sampling patients with sufficient electronic health record data for research. BMC Med Inform Decis Mak. 2014;14:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin JH, Haug PJ. Exploiting missing clinical data in Bayesian network modeling for predicting medical problems. J Biomed Inform. 2008;41(1):1–14. [DOI] [PubMed] [Google Scholar]
- 13.Hripcsak G, Albers DJ, Perotte A. Exploiting time in electronic health record correlations. J Am Med Inform Assoc. 2011;18 Suppl 1:i109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasko TA, Denny JC, Levy MA. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PLoS One. 2013;8(6):e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pivovarov R, Albers DJ, Sepulveda JL, Elhadad N. Identifying and mitigating biases in EHR laboratory tests. J Biomed Inform. 2014;51:24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spratt SE, Batch BC, Davis LP, Dunham AA, Easterling M, Feinglos MN, et al. Methods and initial findings from the Durham Diabetes Coalition: Integrating geospatial health technology and community interventions to reduce death and disability. Journal of Clinical & Translational Endocrinology. 2(1):26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.PCORnet Common Data Model. http://www.pcornet.org/resource-center/pcornet-common-data-model/. Accessed December 16, 2016.
- 18.Kelly K, Grau-Sepulveda MV, Goldstein BA, Spratt SE, Wolfley A, Hatfield V, et al. The agreement of patient-reported versus observed medication adherence in type 2 diabetes mellitus (T2DM). BMJ Open Diabetes Res Care. 2016;4(1):e000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Califf RM, Harrell FE, Jr., Lee KL, Rankin JS, Hlatky MA, Mark DB, et al. The evolution of medical and surgical therapy for coronary artery disease. A 15-year perspective. JAMA. 1989;261(14):2077–86. [PubMed] [Google Scholar]
- 20.Eisenstein EL, Shaw LK, Anstrom KJ, Nelson CL, Hakim Z, Hasselblad V, et al. Assessing the clinical and economic burden of coronary artery disease: 1986-1998. Med Care. 2001;39(8):824–35. [DOI] [PubMed] [Google Scholar]
- 21.Rothwell PM. External validity of randomised controlled trials: “to whom do the results of this trial apply?”. Lancet. 2005;365(9453):82–93. [DOI] [PubMed] [Google Scholar]
- 22.Granger BB, Staton M, Peterson L, Rusincovitch SA. Prevalence and Access of Secondary Source Medication Data: Evaluation of the Southeastern Diabetes Initiative (SEDI). AMIA Jt Summits Transl Sci Proc. 2015;2015:66–70. [PMC free article] [PubMed] [Google Scholar]
- 23.Pitts SR, Adams RP. Emergency department hypertension and regression to the mean. Ann Emerg Med. 1998;31(2):214–8. [DOI] [PubMed] [Google Scholar]
- 24.Stratton IM, Adler AI, Neil HA, Matthews DR, Manley SE, Cull CA, et al. Association of glycaemia with macrovascular and microvascular complications of type 2 diabetes (UKPDS 35): prospective observational study. BMJ. 2000;321(7258):405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiou S-J, Campbell C, Horswell R, Myers L, Culbertson R. Use of the emergency department for less-urgent care among type 2 diabetics under a disease management program. BMC Health Services Research. 2009;9(1):223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Byrne MM, Kuebeler M, Pietz K, Petersen LA. Effect of using information from only one system for dually eligible health care users. Med Care. 2006;44(8):768–73. [DOI] [PubMed] [Google Scholar]
- 27.Richesson RL, Rusincovitch SA, Wixted D, Batch BC, Feinglos MN, Miranda ML, et al. A comparison of phenotype definitions for diabetes mellitus. J Am Med Inform Assoc. 2013;20(e2):e319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spratt SE, Pereira K, Granger BB, Batch BC, Phelan M, Pencina M, et al. Assessing electronic health record phenotypes against gold-standard diagnostic criteria for diabetes mellitus. J Am Med Inform Assoc. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quan H, Sundararajan V, Halfon P, Fong A, Burnand B, Luthi JC, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care. 2005;43(11):1130–9. [DOI] [PubMed] [Google Scholar]
- 30.Balding DJ, Bishop MJ, Cannings C. Handbook of statistical genetics Chichester; New York: Wiley; 2001. xxvi, 863 p. p. [Google Scholar]
- 31.Noel PH, Copeland LA, Perrin RA, Lancaster AE, Pugh MJ, Wang CP, et al. VHA Corporate Data Warehouse height and weight data: opportunities and challenges for health services research. J Rehabil Res Dev. 2010;47(8):739–50. [DOI] [PubMed] [Google Scholar]
- 32.Paul DW, Neely NB, Clement M, Riley I, Al-Hegelan M, Phelan M, et al. Development and validation of an electronic medical record (EMR)-based computed phenotype of HIV-1 infection. J Am Med Inform Assoc. 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernan MA, Robins JM. Using Big Data to Emulate a Target Trial When a Randomized Trial Is Not Available. Am J Epidemiol. 2016;183(8):758–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson CA, Kurian AW, Luft HS. Linking Electronic Health Records to Better Understand Breast Cancer Patient Pathways Within and Between Two Health Systems. eGEMs. 2015;3(1):1127. [DOI] [PMC free article] [PubMed] [Google Scholar]


