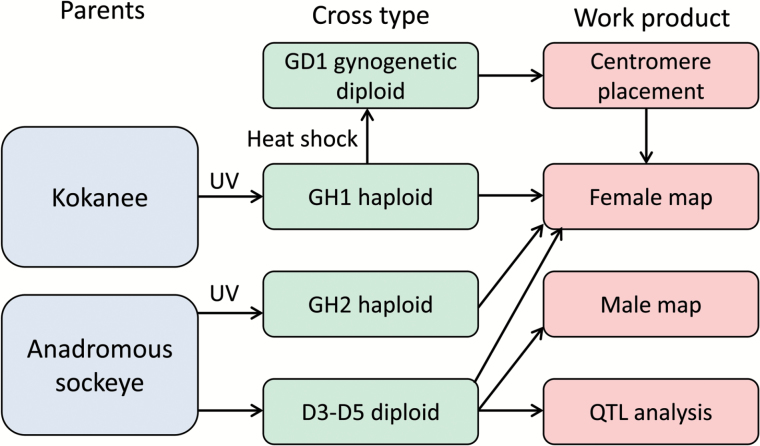
Figure 1.
Workflow for this study. The study included a haploid and gynogenetic diploid family of kokanee (freshwater resident sockeye salmon) sampled at the southern end of the species range and haploid and diploid families of anadromous sockeye salmon sampled at the northern end of the species range (GH2, D3–D5). Gynogenetic haploids (families GH1, GH2) were created by combining eggs and UV irradiated sperm and gynogenetic diploids (family GD1) were created by heat shocking eggs and UV irradiated sperm after fertilization. Diploids (families D3-D5) were created by mating wild individuals from a single population. The female linkage map included data from all 5 families and centromeres were placed on this map using knowledge of recombination events available from the gynogenetic diploids. The male map was constructed using only the diploid families because gynogenetic haploid families do not include information about recombination events in males. QTL analysis for 4 phenotypic traits was conducted for each of the diploid families (D3–D5). Haploid families were not used for QTL analysis because haploid embryos do not survive past hatch. Additional information on each family including sample size can be found in Table 1. Colors are viewable in the online version of the article.