Abstract
The present study determined the expression and biological functions of FOXA2 gene in colon cancer in tissues, cells and animals. A total of 66 patients with colon cancer were included in the present study. Using The Human Protein Atlas database, expression and distribution of FOXA2 in colon cancer tissues were analyzed. Using immunohistochemistry, the expression and distribution of FOXA2 in colon cancer cells were studied. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to determine the expression of FOXA2 mRNA in colon cancer tissues. Following in vitro transfection with FOXA2 interference sequence (siR-FOXA2), the proliferation, cell cycle, migration and invasion of colon cancer HCT116 and HT29 cells were investigated using Cell Counting Kit-8 assay, flow cytometry, and Transwell assay, respectively. Furthermore, flow cytometry was used to determine apoptosis of HCT116 and HT29 cells. Western blotting was used to determine the expression of epithelial mesenchymal transition (EMT) proteins, E-Cadherin and Vimentin. Laser scanning confocal microscopy was performed to observe the cytoskeleton in HCT116 and HT29 cells. Results indicated tumorigenesis of colon cancer cells in nude mice. In addition, the expression of FOXA2 in colon cancer tissues was elevated and associated with the metastasis and clinical staging of colon cancer. Notably, inhibition of FOXA2 reduced the proliferation of colon cancer cells in vitro and reduced expression of FOXA2 was able to decrease the migration and invasion abilities of colon cancer cells. Furthermore, FOXA2 promoted EMT, inhibited apoptosis and enhanced the invasion ability of colon cancer cells. Decreased expression of FOXA2 inhibited tumorigenesis of colon cancer cells in nude mice. To conclude, the present study demonstrated that the expression of FOXA2 in colon cancer tissues was elevated and associated with the metastasis and clinical staging of colon cancer. As an oncogene, FOXA2 may promote the proliferation, migration and invasion and EMT in colon cancer.
Keywords: FOXA2, colon cancer, oncogene, proliferation, migration, invasion, epithelial mesenchymal transition
Introduction
Colon cancer is a clinically common malignant tumor in digestive tract, and it has a very high incidence all over the world (1,2). It is reported that the incidence of colon cancer is the third highest among all malignant tumors (3). At present, surgery combined with radiochemotherapy and immunotherapy are the main ways of clinical treatments for colon cancer (4–6). In patients with early-stage colon cancer, the degree of malignancy is not high, and lymph node metastasis does not occur (7). These patients usually have good postoperative recovery, and a 5-year survival rate of over 60% (8). By contrast, patients with advanced colon cancer usually have poor clinical treatment effects and a 5-year survival rate of 20%, because of widespread metastasis and drug resistance (9). The invasion and metastasis of cancer is the main cause of death in colon cancer patients, but its molecular mechanism is still unclear. Forkhead box (Fox) family genes are abundant in fungi and animal cells, and their main function is to regulate gene expression by binding with DNA promoter region as a transcription factor (10). Fox family is first discovered to regulate embryonic development, and more and more studies show that Fox family proteins play a key regulatory role in immune system, cell cycle, energy metabolism, and cell aging (11,12). FOXA2 gene, a member of Fox family, is localized at human chromosome 20p11, with a length of 45 kb (13,14). It contains 3 exons and 2 introns, and its protein product has 457 amino acids (13,14). The amino acid sequence of its binding site at DNA is 5′-RYMAYAY-3′ (13,14). It is discovered that FOXA2 can bind to the promoter region of genes such as CREB and HNF6, thereby activating their transcription (15,16). Under normal conditions, FOXA2 is expressed differently in liver, lungs, pancreas and other tissues, and its activity is regulated by phosphorylation and acetylation (17). It is also reported that the expression of FOXA2 is abnormal in many tumor tissues, and abnormal FOXA2 is involved in the processes of proliferation, invasion and metastasis, and epithelial mesenchymal transition (EMT) of various tumors, such as liver cancer, prostate cancer, stomach cancer, and bladder cancer (18,19). At present, the expression and function of FOXA2 gene in colon cancer is still unclear.
EMT refers to a transformation process in which epithelial cells, under the influence by certain factors, lose the polarity of epithelial cells and acquire the characteristics of mesenchymal cells (20). This process is accompanied by changes in cell morphology, function, and gene expression profiles (21). EMT is a key step for tumor cells to break away from their original sites and gain the ability of invasion and metastasis, and alleviation of EMT suppresses invasion and metastasis of tumor cells (22). Therefore, it is necessary to study the molecular mechanism of tumor cell EMT. Studies show that a variety of transcription factors play important roles in tumor EMT, including ZEB1, E47 and Twist (23,24). However, whether FOXA2 is involved in the regulation of EMT and thereby affecting tumor metastasis has not been reported before. In the present study, we investigate the expression and mechanism of action of FOXA2 in colon cancer at tissue and cellular levels.
Materials and methods
Patients
A total of 66 patients with colon cancer who received treatments at our hospital between January 2014 and December 2016 were included in the present study. Among the patients, 49 were males, and 17 were females, with an age range of 35–64 years and an average age of 48.5 years. The inclusion criterion was that the patients were initially diagnosed of colon cancer. The exclusion criteria were: i) the patient had other tumors; ii) the patient received radiotherapy or chemotherapy; iii) the patient had a long history of drug intake; iv) the patient had chronic diseases. None of the patients received radiochemotherapy before surgeries. The clinical and pathological data were collected, including tumor size, clinical stage, histological differentiation, lymph node metastasis and distant metastasis. Colon cancer tissues were collected from all patients. In addition, tumor-adjacent tissues were collected as control. All procedures were approved by the Ethics Committee of Qingdao University. Written informed consents were obtained from all patients or their families.
Cells
Normal colonic epithelial NCM460 cells, and colon cancer HCT116 and HT29 cells were cultured in DMEM medium supplemented with 10% fetal bovine serum, 100 IU/ml penicillin and 100 IU/ml streptomycin under 37°C, 5% CO2, and 70% humidity. The cells were passaged every three days, and log-phase cells were collected for experiments.
One day before transfection, log-phase HCT116 and HT29 cells (2×105) were seeded onto 24-well plates, and cultured in serum-free DMEM medium until reaching 70% confluency. In the first vial, 1.5 µl FOXA2 siRNA [20 pmol/µl; FOXA2 interference sequence (siR-FOXA2) group] or 0.5 µg FOXA2 plasmids (Hanbio Biotechnology Co., Ltd., Shanghai, China) was mixed with 50 µl Opti Memi medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA). In the second vial, 1 µl Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) was mixed with 50 µl Opti-MEMI medium. After standing still for 5 min, the two vials were combined for another waiting at room temperature for 20 min. Then, the mixtures were added onto cells in respective groups. Six hours later, the medium was replaced with DMEM medium containing 10% fetal bovine serum. After cultivation at 37°C and 5% CO2 for 48 h, the cells were collected for further assays.
Immunohistochemical data analysis
After logging on The Human Protein Atlas database (http://www.proteinatlas.org/), the name of ‘FOXA2’ was searched to query the expression of FOXA2 in colon cancer and normal colon tissues. In human tissues, FOXA2 scores were given by the website. Search on the website showed that FOXA2 protein expression is up-regulated. Therefore, we tested protein and mRNA expression at cellular and tissue levels as shown below. In addition, positive rate and distribution were analyzed.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Colon cancer tissues and control tissues (100 mg) were ground into powder in liquid nitrogen and mixed with 1 ml Trizol (Thermo Fisher Scientific, Inc.) for lysis. Then, total RNA was extracted using phenol chloroform method. The purity of RNA was determined by A260/A280 using ultraviolet spectrophotometry (Nanodrop ND2000; Thermo Fisher Scientific, Inc.). Then, cDNA was obtained by reverse transcription from 1 µg RNA and stored at −20°C. Reverse transcription of was carried out using miScript II RT kit (Qiagen, Hilden, Germany).
For RT-qPCR, miScript SYBR® Green PCR Kit (Qiagen) was used. The RT-qPCR reaction system was composed of 10 µl RT-qPCR-Mix, 0.5 µl upstream primer (5′-CCCCTGAGTTGGCGGTGGT-3′), 0.5 µl downstream primer (5′-TTGCTCACGGAAGAGTAG-3′), 2 µl cDNA and 7 µl ddH2O. The reaction protocol was: Initial denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 1 min and annealing at 60°C for 30 sec.
Western blotting
Cells (1×106) in each group were trypsinized and collected. Then, precooled radio immunoprecipitation assay (RIPA) lysis buffer (1,000 µl; 50 mM Tris-base, 1 mM EDTA, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Triton X-100, 1% sodium deoxycholate; Beyotime Institute of Biotechnology, Shanghai, China) was added to the samples. After lysis for 30 min on ice, the mixture was centrifuged at 12,000 × g ta 4°C for 10 min. The supernatant was used to determine protein concentration by bicinchoninic acid (BCA) protein concentration determination kit (RTP7102; Real-Times Biotechnology Co., Ltd., Beijing, China). Protein samples (6 µg) were then mixed with 5X sodium dodecyl sulfate loading buffer before denaturation in boiling water bath for 10 min. Afterwards, the samples (10 µl) were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (80 V). The resolved proteins were transferred to polyvinylidene difluoride membranes on ice (100 V, 1 h) and blocked with 50 g/l skimmed milk at room temperature for 1 h. Then, the membranes were incubated with rabbit anti-human FOXA2 polyclonal primary antibody (1:1,000; Abcam, Cambridge, UK), rabbit anti-human E-Cadherin polyclonal primary antibody (1:1,000; Abcam), rabbit anti-human Vimentin polyclonal primary antibody (1:1,000; Abcam), and rabbit anti-human GAPDH primary antibody (1:4,000; Abcam) at 4°C overnight. After extensive washing with phosphate-buffered saline with Tween-20 for 5 times of 5 min, the membranes were incubated with goat anti-rabbit (1:2,000 for FOXA2, E-Cadherin and Vimentin) or goat anti-mouse (1:4,000 for GAPDH)-labelled horseradish peroxidase-conjugated secondary antibodies (Abcam) for 1 h at room temperature before washing with phosphate-buffered saline with Tween-20 for 5 times of 5 min. Then, the membrane was developed with enhanced chemiluminescence detection kit (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for imaging. Image lab v3.0 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to acquire and analyze imaging signals. The relative contents of target proteins were expressed against GAPDH.
Cell Counting Kit-8 (CCK-8) assay
Cells were seeded at a density of 2,000/well in 96-well plates. At 0, 24, 48 and 72 h, 20 µl CCK-8 (5 g/l) solution (Beyotime Institute of Biotechnology) was added onto the cells. On the last day, 150 µl CCK-8 reaction solution was added and the cells were incubated at 37°C for 2 h. The absorbance of each well was measured at 490 nm for plotting cell proliferation curves. Each group was tested in 3 replicate wells and the values were averaged.
Transwell assay
Matrigel was thawed at 4°C overnight and diluted with serum-free DMEM medium (dilution 1:3). The mixture (50 µl) was evenly smeared into the upper chamber (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and incubated at 37°C for 1 h. After solidification, 1×105 cells from each group were seeded into the upper chamber containing 200 µl serum-free DMEM medium. In addition, 500 µl DMEM medium supplemented with 10% fetal bovine serum was added into the lower chamber. After incubation at 37°C and 5% CO2 for 24 h, the chamber was removed and the cells in the upper chamber were wiped off. After being fixed with 4% formaldehyde for 10 min, the membrane was stained using Giemsa method for microscopic observation of 5 random fields (magnification, ×200). The number of transwell cells was calculated for the evaluation of cell invasion and migration ability. All procedures were carried out on ice with pipetting tips being precooled at 4°C.
Flow cytometry
At 24 h after transfection, cells (1×106) in each group were washed with pre-cooled phosphate-buffered saline twice and subjected to cell cycle detection using Cycletest™ Plus DNA Reagent kit (BD Biosciences, Franklin Lakes, NJ, USA) following the manufacturer's manual. The data were analyzed using ModFit software (Verity Software House, Topsham, ME, USA).
Laser scanning confocal microscopy
At 24 h after transfection with siR-FOXA2, cells (1×105) in each group were seeded onto petri-dishes (diameter, 6 cm) and incubated at 37°C and under 5% CO2 for 24 h. After discarding medium, the cells were washed with phosphate-buffered saline for three times, and fixed with 4% formaldehyde for 10 min. After washing with phosphate-buffered saline for 3 times, the cells were stained with 5 µM rhodamine for 5 min. After additional washing with phosphate-buffered saline for 3 times, the cells were visualized under a laser scanning confocal microscope (SP8; Leica, Wetzlar, Germany).
In vivo assay in animal model
Sixteen BALB/C nude mice were divided evenly into two groups, negative control (NC) and siR-FOXA2 group. NC and siR-FOXA2 sequences were transfected into HCT116 cells using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) following the manufacturer's manual. At 24 h after transfection, the cells (2×106) in each group were trypsinized and resuspended with sterile phosphate-buffered saline (0.2 ml). Then, the cell suspension was inoculated in nude mouse armpit to construct xenograft tumor model nude mice. Vital signs of nude mice were observed, and the mice were sacrificed in week 5 to extract tumor tissues. After fixation with 4% formaldehyde, the tumor tissues were dehydrated, paraffin-embedded, and sliced (4 nm thick) for immunohistochemistry test. E-Cadeherin and Vimentin rabbit anti-human polyclonal antibodies (Beyotime Institute of Biotechnology) were diluted at a ratio of 1:50 with water, and used to incubate the slices at 4°C overnight. After washing with phosphate-buffered saline twice, goat anti-rabbit secondary antibody was added, followed by incubation at 37°C for 30 min before color development. All animal experiments were conducted according to the ethical guidelines of Qingdao University (Qingdao, China).
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (IBM Corp., Armonk, NY, USA). The data were expressed as mean ± standard deviation and intergroup comparison was carried out using Student's t-test. Comparison of multiple groups was performed using ANOVA followed by Tukey's or Dunnett's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of FOXA2 in colon cancer tissues is elevated, and closely related with the metastasis and clinical staging of colon cancer
To examine the localization of FOXA2, we used immunohistochemistry. By comparing with public immunohistochemical database (http://www.proteinatlas.org/), we discovered that FOXA2 expression in colon cancer tissues was significantly higher than that in normal colon tissues (all data from Protein Atlas database; Fig. 1A). Immunohistochemical data showed that FOXA2 was localized in the nucleus of NCM460 cells, and the nucleus, cytoplasm and membrane of HCT116 and HT29 cells (Fig. 1B). To measure the expression of FOXA2, RT-qPCR was performed. The data showed that FOXA2 expression in colon cancer tissues was significantly higher than that in normal colon tissues (Fig. 1C). In addition, FOXA2 expression in colon cancer tissues from patients with lymphatic metastasis was significantly higher than that from patients without lymphatic metastasis (Fig. 1D). Clinical staging showed that FOXA2 level in colon cancer tissues at IV stage was significantly higher than those at I, II and III stages (Fig. 1E). These results suggest that the expression of FOXA2 in colon cancer tissues is elevated, and closely related with the metastasis and clinical staging of colon cancer.
Figure 1.
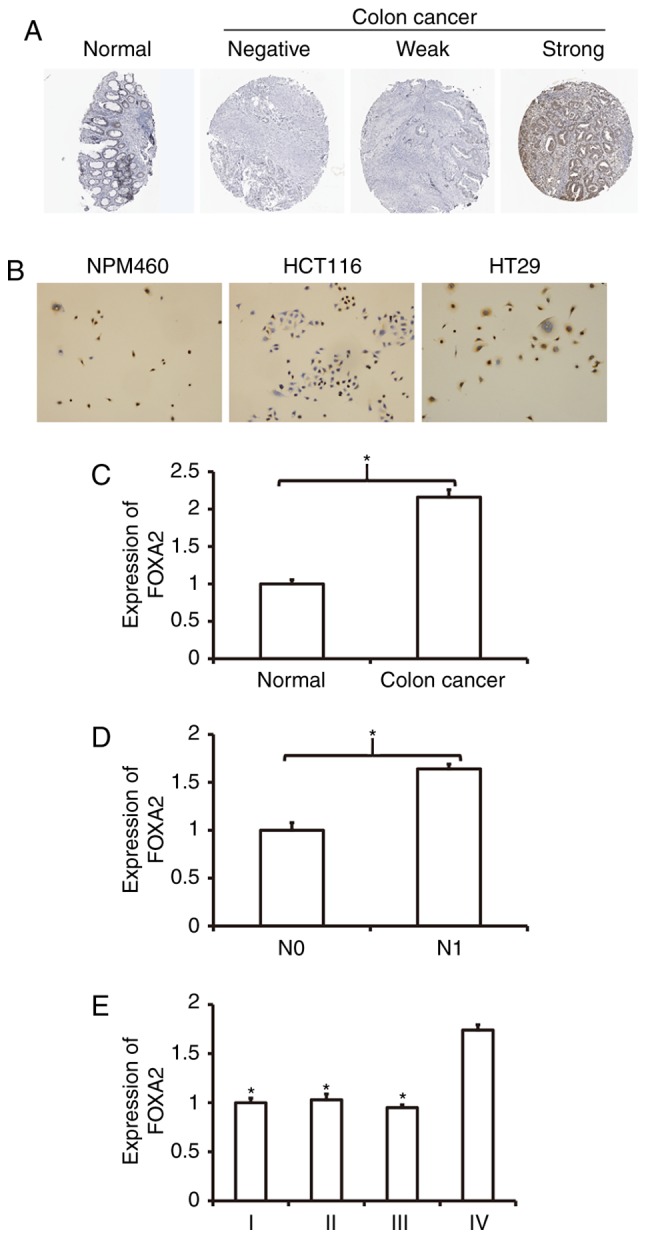
Expression and distribution of FOXA2 in colon cancer. (A) Expression of FOXA2 in colon cancer tissues shown by immunohistochemistry. The figure was obtained from the Human Protein Atlas database (https://www.proteinatlas.org/ENSG00000125798-FOXA2/tissue/colon#) (38). (B) Expression and distribution of FOXA2 in colon cancer HCT116 and HT29 cells. Magnification, ×100. (C-E) Expression of FOXA2 mRNA in (C) normal and colon cancer tissues (*P<0.05 compared with normal), (D) patients without/with lymphatic metastasis (*P<0.05 compared with stage N0), or (E) patients in different clinical stages (comparison of multiple groups was performed using ANOVA followed by Tukey's/Dunnett's test; *P<0.05 compared with IV).
Inhibition of FOXA2 reduces the proliferation of colon cancer cells in vitro
To determine the expression of FOXA2 protein, western blotting was used. The data showed that transfection with siR-FOXA2 reduced the protein expression of FOXA2 in HCT116 and HT29 cells (Fig. 2A). To detect the proliferation of HCT116 and HT29 cells, CCK-8 assay was carried out. The data showed that the proliferation of HCT116 and HT29 cells transfected with siR-FOXA2 was significantly reduced compared with negative control group (P<0.05; Fig. 2B). Flow cytometry showed that G1/S phase transition in HCT116 and HT29 cells transfected with siR-FOXA2 was reduced compared with negative control group (Fig. 2C). These results indicate that inhibition of FOXA2 reduces the proliferation of colon cancer cells in vitro, and suppresses the transition from G1 phase to S phase.
Figure 2.
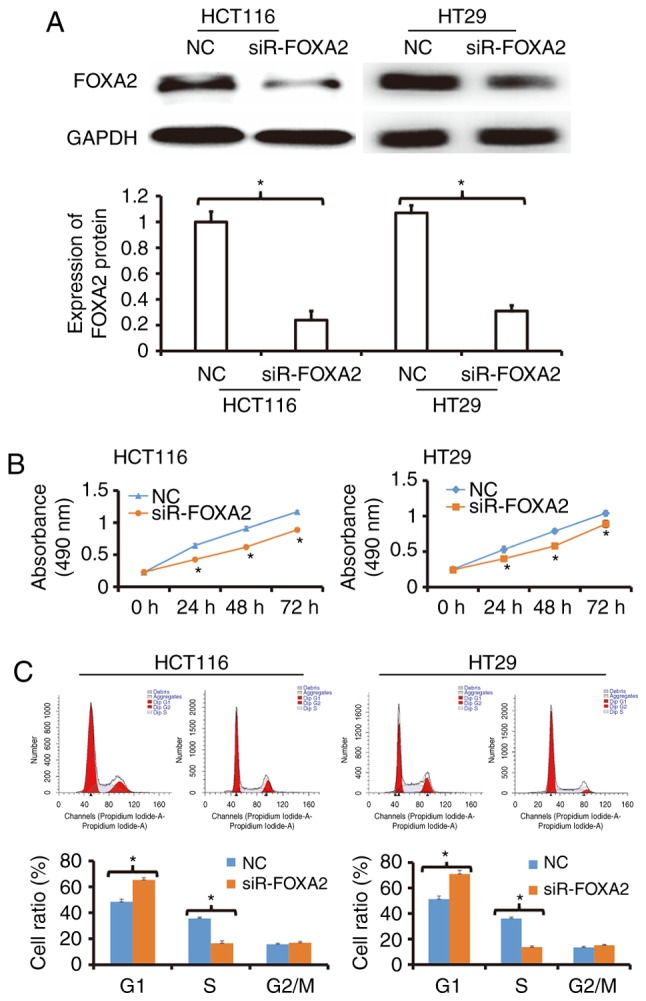
Effect of FOXA2 on the proliferation of colon cancer cells in vitro. (A) Expression of FOXA2 in HCT116 and HT29 cells transfected with FOXA2 interference sequence (siR-FOXA2). Western blotting was used to determine protein expression. *P<0.05 compared with negative control (NC) as indicated. (B) Proliferation curves for HCT116 and HT29 cells transfected with siR-FOXA2. CCK-8 assay was performed to detect cell proliferation. *P<0.05 compared with NC. (C) Percentages of cells in G1, S or G2/M phases. Flow cytometry was used to detect cell phases. *P<0.05 compared with NC as indicated.
Reduced expression of FOXA2 is able to decrease the migration and invasion abilities of colon cancer cells
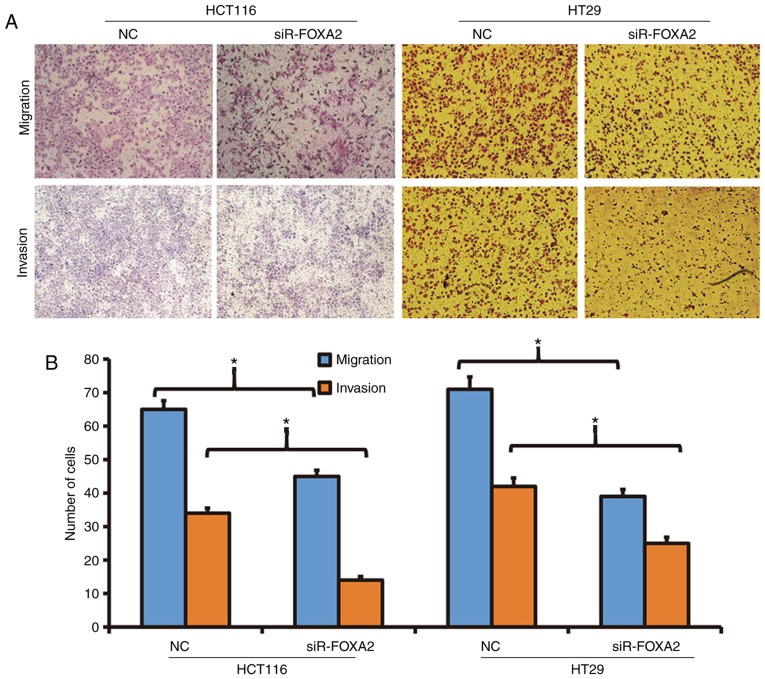
To determine the migration and invasion abilities of HCT116 and HT29 cells, Transwell assay was employed. The data showed that the number of transwell HCT116 cells after transfection with siR-FOXA2 was significantly reduced than that in negative control group in both migration and invasion assays (P<0.05; Fig. 3). Similarly, the number of transwell HT29 cells after transfection with siR-FOXA2 was significantly lower than that in negative control group in both migration and invasion assays (P<0.05; Fig. 3). The results suggest that reduced expression of FOXA2 is able to decrease the migration and invasion abilities of colon cancer cells.
Figure 3.
Effect of FOXA2 on the migration and invasion of colon cancer cells. (A) Images of HCT116 and HT29 cells transfected with FOXA2 interference sequence (siR-FOXA2). Magnification, ×100. (B) Number of HCT116 and HT29 cells transfected with siR-FOXA2. Transwell assay was performed to determine migration and invasion abilities. *P<0.05 compared with NC as indicated.
FOXA2 may promote EMT, inhibit apoptosis, and enhance the invasion ability of colon cancer cells
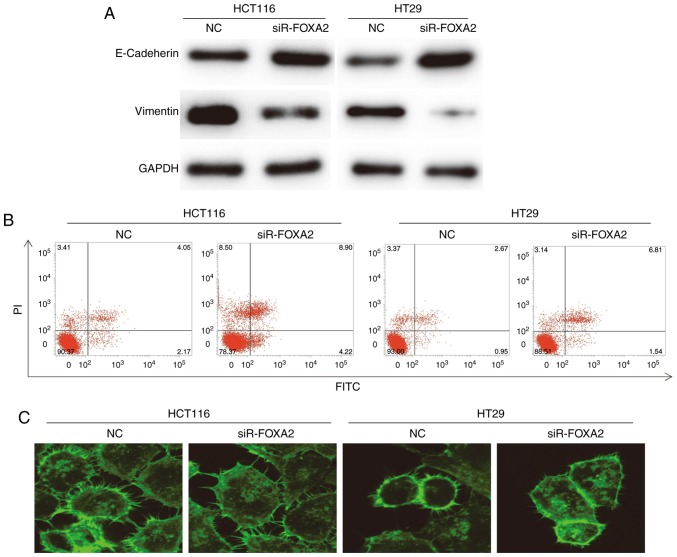
To detect EMT, apoptosis and cytoskeleton, Western blotting, flow cytometry and laser scanning confocal microscopy were carried out, respectively. Western blots showed that the expression of E-Cadherin in HCT116 and HT29 cells transfected with siR-FOXA2 was significantly higher than that in NC group, while expression of Vimentin in HCT116 and HT29 cells transfected with siR-FOXA2 was lower than that in NC group (Fig. 4A). In the meantime, flow cytometry showed that the apoptosis of HCT116 and HT29 cells transfected with siR-FOXA2 was enhanced than those of NC groups (Fig. 4B). Laser scanning confocal microscopy showed that protrusion of the cell membrane in HCT116 and HT29 cells transfected with siR-FOXA2 was reduced than that in NC group (Fig. 4C). These results indicate that FOXA2 may promote EMT, inhibit apoptosis, and enhance the invasion ability of colon cancer cells.
Figure 4.
Effect of FOXA2 on epithelial mesenchymal transition of colon cancer cells. (A) Expression of E-Cadherin and Vimentin in HCT116 and HT29 cells transfected with FOXA2 interference sequence (siR-FOXA2). Western blotting was used to measure the expression of E-Cadherin and Vimentin. (B) Apoptosis of HCT116 and HT29 cells transfected with siR-FOXA2. Flow cytometry was performed to detect apoptosis. (C) Protrusion of the cell membrane in HCT116 and HT29 cells transfected with siR-FOXA2. Laser scanning confocal microscopy was used to observe the cells (magnification, ×400).
Decreased expression of FOXA2 inhibits tumorigenesis of colon cancer cells in nude mice
To test the effect of FOXA2 on the formation of colon tumor, nude mice were used. Tumor diameter in siR-FOXA2 group seemed smaller than that in NC group (Fig. 5A). Similarly, weights of colon tumors from mice inoculated with HCT116 or HT29 cells transfected with siR-FOXA2 were significantly lower than that in NC group, respectively (P<0.05; Fig. 5B). Immunohistochemistry showed that E-Cadherin expression in siR-FOXA2 group seemed higher than that in NC group, while Vimentin expression in in siR-FOXA2 group seemed lower than that in NC group (Fig. 5C). The results suggest that decreased expression of FOXA2 inhibits tumorigenesis of colon cancer cells in nude mice.
Figure 5.
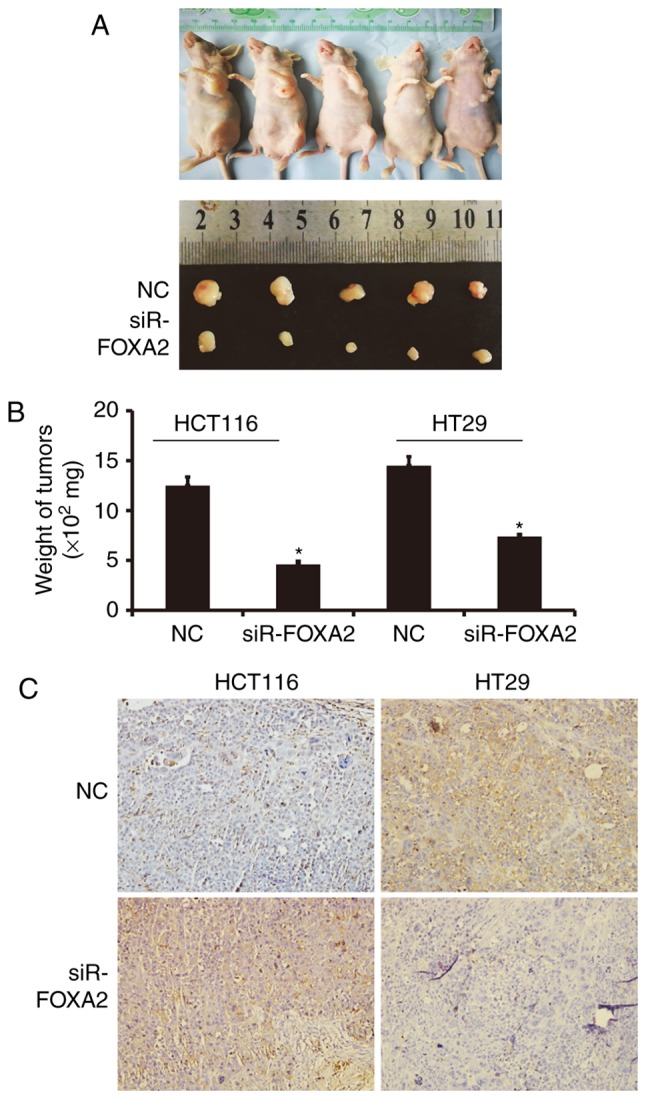
Effect of FOXA2 on tumorigenesis of colon cancer cells in nude mice. (A) Representative images of xenograft formed by HCT116 cells transfected with FOXA2 interference sequence (siR-FOXA2). (B) Weight of tumors from nude mice inoculated with HCT116 or HT29 cells that were transfected with siR-FOXA2. *P<0.05 vs. negative control (NC). (C) Expression of E-Cadherin and Vimentin in xenograft tissues derived from HCT116 cells that were transfected with siR-FOXA2. Immunohistochemistry was used to detect the expression of E-Cadherin and Vimentin. Magnification, ×200.
Discussion
Tumor recurrence and metastasis are the key factors that restrict the postoperative survival of colon cancer patients (25). Studies show that the recurrence and metastasis of colon cancer are closely related to gene expression, methylation, mutation, tumor stem cells, drug resistance and immunosuppression (7,26). Transcription factors play key regulatory roles in the process of gene expression regulation, and they are important drug targets (27). In the present study, we discover that the expression of FOXA2 is up-regulated in colon cancer tissues, and correlated with lymphatic metastasis and clinical staging of colon cancer. In vitro and in vivo experiments show that FOXA2 can promote the proliferation, migration and invasion of colon cancer cells, and EMT may be the key factor for FOXA2 to promote metastasis of colon cancer. These results suggest that FOXA2 plays an oncogene role by promoting EMT and colon cancer recurrence and metastasis.
Transcription factors can bind to the binding sites of the promoter region of genes, and initiate gene transcription, having important regulatory functions (28,29). In cancer, transcription factors are often able to initiate transcription of multiple downstream oncogenes or tumor-suppressor genes, thereby inducing a cascade of responses that regulate cell proliferation, invasion, metastasis, differentiation, drug resistance, and apoptosis (30,31). Studies show that multiple transcription factors are closely related to the recurrence and metastasis of colon cancer. For example, Ma et al discover that KLF4 inhibits the proliferation of colon cancer by activating NDRG2 expression (32). Stein et al discover that MACC1 indirectly activates the HGF/c-Met signaling pathway by activating transcription of c-Met gene, and promotes the recurrence and metastasis of colon cancer (33). In the present study, we find that transcription factor FOXA2 is up-regulated in colon cancer tissues. Moreover, our immunohistochemical data show that FOXA2 expression is distributed in the nucleus, cytoplasm and membrane in colon cancer, which is abnormal. Our RT-qPCR data demonstrate that expression of FOXA2 is up-regulated in colon cancer tissues, and positively correlated with lymphatic metastasis and clinical staging. These results indicate that FOXA2 is associated with the development and progression of colon cancer and may play the role of an oncogene.
Some studies show that the function of FOXA2 in tumors is two-sided. For example, Li et al report that miR-187 can target the expression of FOXA2 and promote the proliferation and metastasis of gastric cancer, suggesting that FOXA2 may be a tumor-suppressor gene in gastric cancer (34). Down-regulation of FOXA2 may promote EMT in pancreatic cancer, suggesting that FOXA2 may inhibit tumor EMT (35). Tu et al discover that miR-1291 inhibits proliferation and metastasis of pancreatic cancer by targeting FOXA2 (36). Our results in the present study show that FOXA2 expression is elevated in HCT116 and HT29 cells. CCK-8 assay shows that interference of FOXA2 expression reduces the proliferation of HCT116 and HT29 cells. In addition, HCT116 and HT29 cells in FOXA2 interference group have shown G1/S arrest, suggesting that down-regulation of FOXA2 inhibits the proliferation of colon cancer cells. Transwell assay shows that the number of cells that cross the membrane in FOXA2 interference group is lower than that in control group, suggesting that FOXA2 is able to promote the migration and invasion of colon cancer cells. Furthermore, our Western blotting data show that expression of E-Cadherin and Vimentin in FOXA2 interference group is up-regulated and down-regulated, respectively than that in control group, suggesting that EMT of colon cancer is inhibited by down-regulation of FOXA2. Of note, the apoptosis of cells in FOXA2 interference group is enhanced than control group, and laser scanning confocal microscopy shows that the cytoskeleton in the cell membrane of the FOXA2 interference group is more abundant, suggesting that these cells have stronger mobility. Tumor formation in nude mice shows that the growth rate of tumor in FOXA2 interference group is significantly lower than that in NC group. In addition, E-Cadherin expression is up-regulated and Vimentin expression is down-regulated in tumor tissues of FOXA2 interference group, suggesting that EMT is inhibited. These results indicate that FOXA2 promotes the proliferation, migration and EMT of colon cancer cells.
In conclusion, the present study demonstrates that FOXA2 is an oncogene in colon cancer, and the up-regulation of its expression promotes the proliferation, migration and invasion of colon cancer. As a transcription factor, up-regulation of FOXA2 expression has clinical values in the diagnosis and prognosis evaluation of colon cancer. Because FOXA2 initiates the expression of a large number of downstream genes, it can also be a potential diagnostic and therapeutic target for colon cancer, being consistent with a previous report (37). In future studies, we will investigate the molecular mechanisms of FOXA2 in colon cancer, such as downstream transcriptional genes of FOXA2 and relevant signaling pathways.
Acknowledgements
The authors wish to thank their department and research team for their help and dedication.
Funding
The present work was supported by the National Natural Science Foundation of China (grant no. 81172031).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
BW and YL designed the study; BW, GL, LD and JZ were involved in designing and performing experiments; BW, GL and YL analyzed the data. The final version of the manuscript has been read and approved by all authors, and each author believes that the manuscript represents honest work.
Ethics approval and consent to participate
All procedures performed in the current study were approved by the Ethics Committee of Qingdao University. Written informed consent was obtained from all patients or their families.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chen Y, Fang L, Li G, Zhang J, Li C, Ma M, Guan C, Bai F, Lyu J, Meng QH. Synergistic inhibition of colon cancer growth by the combination of methylglyoxal and silencing of glyoxalase I mediated by the STAT1 pathway. Oncotarget. 2017;8:54838–54857. doi: 10.18632/oncotarget.18601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chino XMS, Martinez CJ, Garzón VRV, González IÁ, Treviño SV, Bujaidar EM, Ortiz GD, Hoyos RB. Cooked chickpea consumption inhibits colon carcinogenesis in mice induced with azoxymethane and dextran sulfate sodium. J Am Coll Nutr. 2017;36:391–398. doi: 10.1080/07315724.2017.1297744. [DOI] [PubMed] [Google Scholar]
- 3.Cusimano A, Balasus D, Azzolina A, Augello G, Emma MR, Di Sano C, Gramignoli R, Strom SC, McCubrey JA, Montalto G, Cervello M. Oleocanthal exerts antitumor effects on human liver and colon cancer cells through ROS generation. Int J Oncol. 2017;51:533–544. doi: 10.3892/ijo.2017.4049. [DOI] [PubMed] [Google Scholar]
- 4.Cho N, Ransom TT, Sigmund J, Tran T, Cichewicz RH, Goetz M, Beutler JA. Growth inhibition of colon cancer and melanoma cells by versiol derivatives from a paraconiothyrium species. J Nat Prod. 2017;80:2037–2044. doi: 10.1021/acs.jnatprod.7b00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myint ZW, Goel G. Role of modern immunotherapy in gastrointestinal malignancies: A review of current clinical progress. J Hematol Oncol. 2017;10:86. doi: 10.1186/s13045-017-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goel G, Sun W. Advances in the management of gastrointestinal cancers-an upcoming role of immune checkpoint blockade. J Hematol Oncol. 2015;8:86. doi: 10.1186/s13045-015-0185-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou PC, Li YH, Lin SC, Lin SC, Lee JC, Lin BW, Liou JP, Chang JY, Kuo CC, Liu YM, et al. Hypoxia-induced downregulation of DUSP-2 phosphatase drives colon cancer stemness. Cancer Res. 2017;77:4305–4316. doi: 10.1158/0008-5472.CAN-16-2990. [DOI] [PubMed] [Google Scholar]
- 8.Pahwa M, Harris MA, MacLeod J, Tjepkema M, Peters PA, Demers PA. Sedentary work and the risks of colon and rectal cancer by anatomical sub-site in the Canadian census health and environment cohort (CanCHEC) Cancer Epidemiol. 2017;49:144–151. doi: 10.1016/j.canep.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, et al. Differentiation of human pluripotent stem cells into colonic organoids via transient activation of BMP signaling. Cell Stem Cell. 2017;21:51–64.e6. doi: 10.1016/j.stem.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ancey PB, Ecsedi S, Lambert MP, Talukdar FR, Cros MP, Glaise D, Narvaez DM, Chauvet V, Herceg Z, Corlu A, Hernandez-Vargas H. TET-catalyzed 5-hydroxymethylation precedes HNF4A promoter choice during differentiation of bipotent liver progenitors. Stem Cell Reports. 2017;9:264–278. doi: 10.1016/j.stemcr.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Von Stetina SE, Liang J, Marnellos G, Mango SE. Temporal regulation of epithelium formation mediated by FoxA, MKLP1, MgcRacGAP, and PAR-6. Mol Biol Cell. 2017;28:2042–2065. doi: 10.1091/mbc.e16-09-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McFadden VC, Shalaby RE, Iram S, Oropeza CE, Landolfi JA, Lyubimov AV, Maienschein-Cline M, Green SJ, Kaestner KH, McLachlan A. Hepatic deficiency of the pioneer transcription factor FoxA restricts hepatitis B virus biosynthesis by the developmental regulation of viral DNA methylation. PLoS Pathog. 2017;13:e1006239. doi: 10.1371/journal.ppat.1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Machado Dantas AC, Guo M, Sagendorf JM, Zhou Z, Jiang L, Chen X, Wu D, Qu L, Chen Z, et al. Structure of the forkhead domain of FOXA2 bound to a complete DNA consensus site. Biochemistry. 2017;56:3745–3753. doi: 10.1021/acs.biochem.7b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bastidas-Ponce A, Roscioni SS, Burtscher I, Bader E, Sterr M, Bakhti M, Lickert H. Foxa2 and Pdx1 cooperatively regulate postnatal maturation of pancreatic β-cells. Mol Metab. 2017;6:524–534. doi: 10.1016/j.molmet.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausa FM, III, Hughes DE, Costa RH. Stability of the hepatocyte nuclear factor 6 transcription factor requires acetylation by the CREB-binding protein coactivator. J Biol Chem. 2004;279:43070–43076. doi: 10.1074/jbc.M407472200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Rubins NE, Ahima RS, Greenbaum LE, Kaestner KH. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab. 2005;2:141–148. doi: 10.1016/j.cmet.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Lehner F, Kulik U, Klempnauer J, Borlak J. Inhibition of the liver enriched protein FOXA2 recovers HNF6 activity in human colon carcinoma and liver hepatoma cells. PLoS One. 2010;5:e13344. doi: 10.1371/journal.pone.0013344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg DT, Gerlitz B, Sharma GR, Richardson MA, Stephens EJ, Grubbs RL, Holmes KC, Fynboe K, Montani D, Cramer MS, et al. FoxA2 involvement in suppression of protein C, an outcome predictor in experimental sepsis. Clin Vaccine Immunol. 2006;13:426–432. doi: 10.1128/CVI.13.3.426-432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanaki M, Tiniakou I, Thymiakou E, Kardassis D. Physical and functional interactions between nuclear receptor LXRα and the forkhead box transcription factor FOXA2 regulate the response of the human lipoprotein lipase gene to oxysterols in hepatic cells. Biochim Biophys Acta. 2017;1860:848–860. doi: 10.1016/j.bbagrm.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Li D, He C, Wang J, Wang Y, Bu J, Kong X, Sun D. MicroRNA-138 inhibits cell growth, invasion and EMT of non-small cell lung cancer via SOX4/p53 feedback loop. Oncol Res. 2017 Jun 13; doi: 10.3727/096504017X14973124850905. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Q, Dai J, Chen T, Dada LA, Zhang X, Zhang W, DeCamp MM, Winn RA, Sznajder JI, Zhou G. Downregulation of PKCζ/Pard3/Pard6b is responsible for lung adenocarcinoma cell EMT and invasion. Cell Signal. 2017;38:49–59. doi: 10.1016/j.cellsig.2017.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahl GM, Spike BT. Cell state plasticity, stem cells, EMT, and the generation of intra-tumoral heterogeneity. NPJ Breast Cancer. 2017;3:14. doi: 10.1038/s41523-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Z, Zheng X, Cao L, Liang N. MicroRNA-539 inhibits the epithelial-mesenchymal transition of esophageal cancer cells by twist-related protein 1-mediated modulation of melanoma associated antigen A4 (MAGEA4) Oncol Res. 2017 Jun 12; doi: 10.3727/096504017X14972679378357. (Epub Ahead of Print) [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Lazarova D, Bordonaro M. ZEB1 mediates drug resistance and EMT in p300-deficient CRC. J Cancer. 2017;8:1453–1459. doi: 10.7150/jca.18762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aquino RGF, Vasques PHD, Cavalcante DIM, Oliveira ALS, Oliveira BMK, Pinheiro LGP. Invasive ductal carcinoma: Relationship between pathological characteristics and the presence of axillary metastasis in 220 cases. Rev Col Bras Cir. 2017;44:163–170. doi: 10.1590/0100-69912017002010. [DOI] [PubMed] [Google Scholar]
- 26.Laudato S, Patil N, Abba ML, Leupold JH, Benner A, Gaiser T, Marx A, Allgayer H. P53-induced miR-30e-5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int J Cancer. 2017;141:1879–1890. doi: 10.1002/ijc.30854. [DOI] [PubMed] [Google Scholar]
- 27.Schayek H, Laitman Y, Katz LH, Pras E, Ries-Levavi L, Barak F, Friedman E. Colorectal and endometrial cancer risk and age at diagnosis in BLMAsh mutation carriers. Isr Med Assoc J. 2017;19:365–367. [PubMed] [Google Scholar]
- 28.Liu Z, Zhao Y, Fang J, Cui R, Xiao Y, Xu Q. SHP2 negatively regulates HLA-ABC and PD-L1 expression via STAT1 phosphorylation in prostate cancer cells. Oncotarget. 2017;8:53518–53530. doi: 10.18632/oncotarget.18591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson AC, Cutty SJ, Gasiunas SN, Deplae I, Stemple DL, Wardle FC. In vivo regulation of the zebrafish endoderm progenitor niche by T-Box transcription factors. Cell Rep. 2017;19:2782–2795. doi: 10.1016/j.celrep.2017.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y, Nar R, Fan AX, Aryan M, Hossain MA, Gurumurthy A, Wassel PC, Tang M, Lu J, Strouboulis J, Bungert J. Functional interrelationship between TFII-I and E2F transcription factors at specific cell cycle gene loci. J Cell Biochem. 2018;119:712–722. doi: 10.1002/jcb.26235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo HS, Ku JM, Choi HS, Woo JK, Lee BH, Kim DS, Song HJ, Jang BH, Shin YC, Ko SG. Apigenin overcomes drug resistance by blocking the signal transducer and activator of transcription 3 signaling in breast cancer cells. Oncol Rep. 2017;38:715–724. doi: 10.3892/or.2017.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y, Wu L, Liu X, Xu Y, Shi W, Liang Y, Yao L, Zheng J, Zhang J. KLF4 inhibits colorectal cancer cell proliferation dependent on NDRG2 signaling. Oncol Rep. 2017;38:975–984. doi: 10.3892/or.2017.5736. [DOI] [PubMed] [Google Scholar]
- 33.Stein U, Walther W, Arlt F, Schwabe H, Smith J, Fichtner I, Birchmeier W, Schlag PM. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15:59–67. doi: 10.1038/nm.1889. [DOI] [PubMed] [Google Scholar]
- 34.Li C, Lu S, Shi Y. MicroRNA-187 promotes growth and metastasis of gastric cancer by inhibiting FOXA2. Oncol Rep. 2017;37:1747–1755. doi: 10.3892/or.2017.5370. [DOI] [PubMed] [Google Scholar]
- 35.Kondratyeva LG, Sveshnikova AA, Grankina EV, Chernov IP, Kopantseva MR, Kopantzev EP, Sverdlov ED. Downregulation of expression of mater genes SOX9, FOXA2, and GATA4 in pancreatic cancer cells stimulated with TGFβ1 epithelial-mesenchymal transition. Dokl Biochem Biophys. 2016;469:257–259. doi: 10.1134/S1607672916040062. [DOI] [PubMed] [Google Scholar]
- 36.Tu MJ, Pan YZ, Qiu JX, Kim EJ, Yu AM. MicroRNA-1291 targets the FOXA2-AGR2 pathway to suppress pancreatic cancer cell proliferation and tumorigenesis. Oncotarget. 2016;7:45547–45561. doi: 10.18632/oncotarget.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayachandran A, Dhungel B, Steel JC. Epithelial-to-mesenchymal plasticity of cancer stem cells: Therapeutic targets in hepatocellular carcinoma. J Hematol Oncol. 2016;9:74. doi: 10.1186/s13045-016-0307-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.