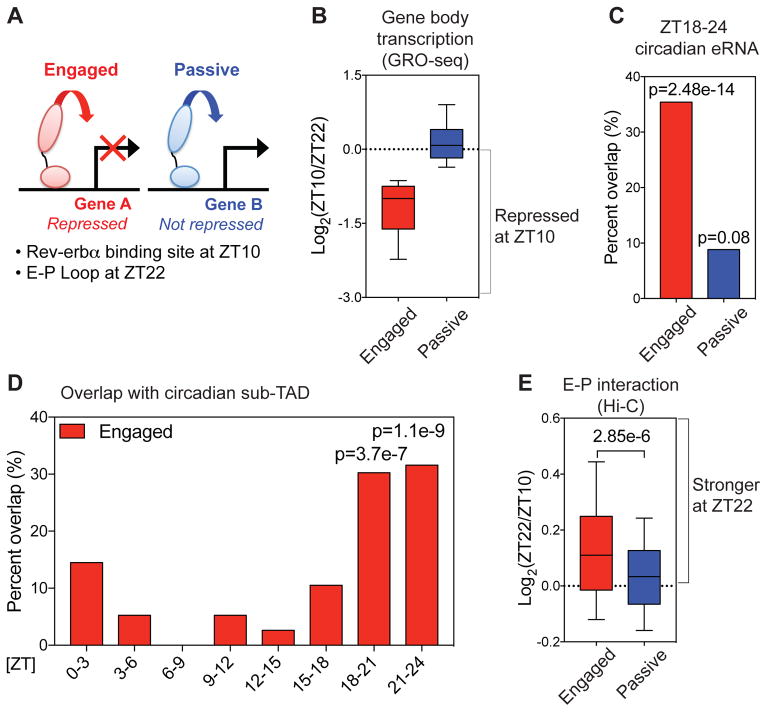
Figure 3. Rev-erbα attenuates enhancer-promoter looping at functional binding sites.
(A) Rev-erbα sites at E-P loops were classified as “engaged” when looped to genes whose transcription was repressed at ZT10 relative to ZT22, and otherwise classified as “passive”. (B) Gene body transcription change between ZT22 and ZT10 at engaged vs. passive sites. Engaged target genes were defined based on nascent gene body transcription fold change ≥1.5 between ZT22 and ZT10. (C) Engaged Rev-erbα sites were highly correlated with ZT18-24 circadian eRNAs (6) (within ± 2kbp, one-tailed hypergeometric tests). (D) Engaged Rev-erbα sites were confined within sub-TADs that contain circadian genes peaking at ZT18-24 (one-tailed hypergeometric tests). (E) E-P loops between engaged Rev-erbα binding sites and target gene promoters were stronger at ZT22 than ZT10 (Mann-Whitney test). For boxplots, whiskers drawn at 10th and 90th percentiles