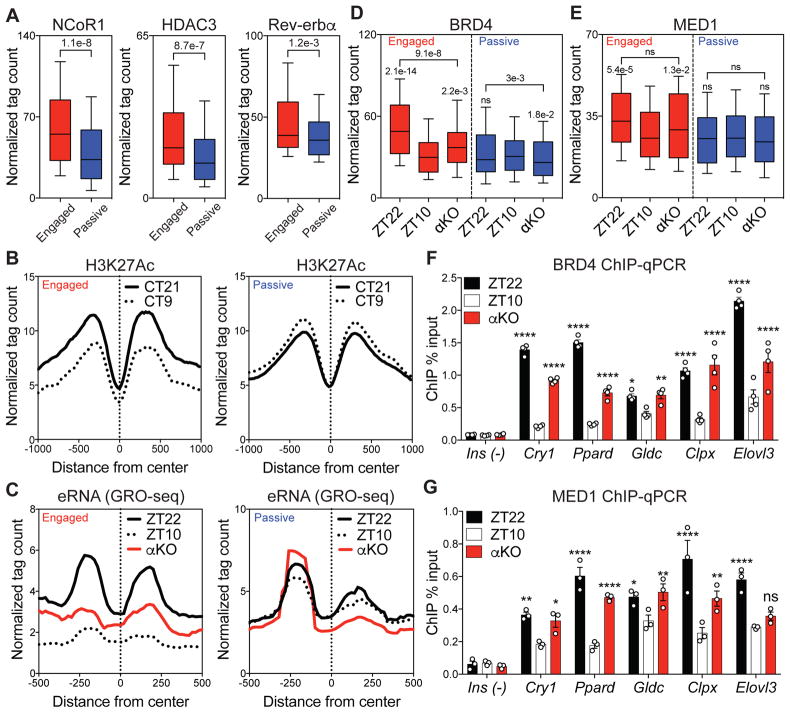
Figure 4. Functional Rev-erbα binding evicts BRD4 and MED1 from sites of looping.
(A) Higher recruitment of NCoR1 and HDAC3 at engaged Rev-erbα sites associated with a slight average increase in Rev-erbα binding (Mann-Whitney tests). (B) Circadian deacetylation of histone 3 lysine 27 (H3K27Ac) at circadian time 21 (CT21, black) and CT9 (dotted) at engaged vs. passive sites. (C) Circadian eRNA transcription between ZT22 (black) and ZT10 (dotted), with increased transcription at ZT10 in αKO (red) at engaged sites. (D) Circadian eviction of BRD4 and (E) MED1 between ZT22 and ZT10, with enhanced binding at ZT10 in αKO at engaged sites (Dunn’s multiple comparisons tests after one-way ANOVA/Friedman test). (F) ChIP-qPCR validation of BRD4 and (G) MED1 eviction at ZT10 and enhanced binding at ZT10 in αKO at engaged sites (Ins as a negative control, n=3–4, two-way ANOVA followed by Dunnett’s multiple comparisons test). For boxplots, whiskers drawn at 10th and 90th percentiles, ns p≥0.05, * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001