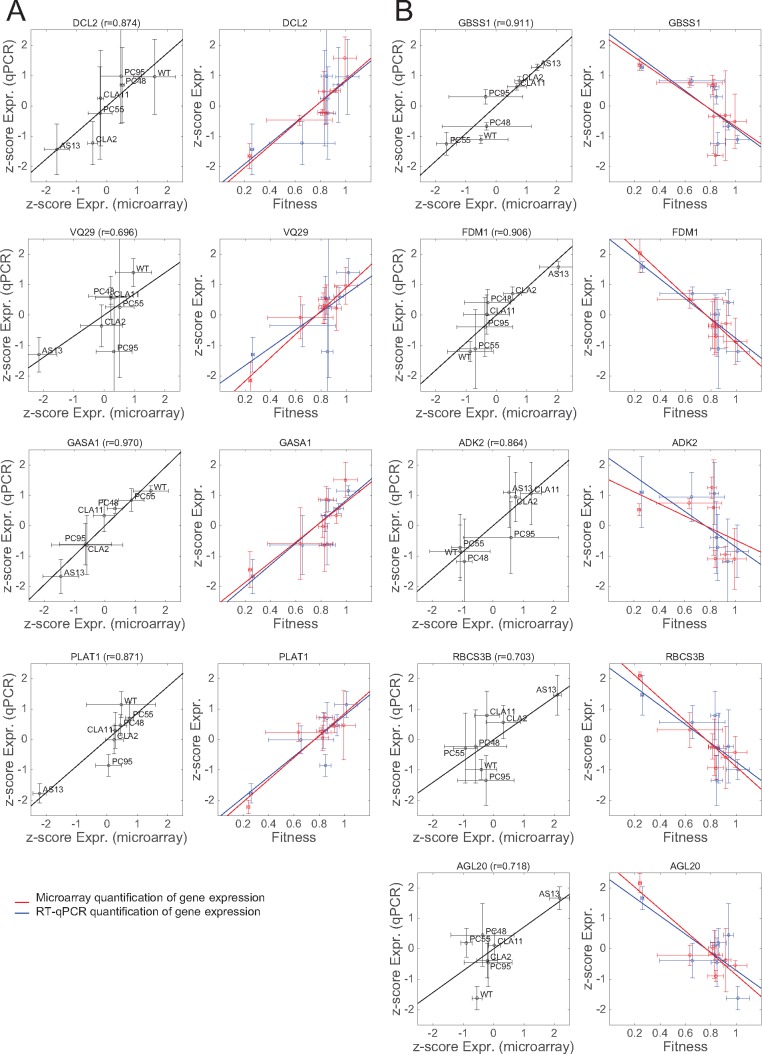
Fig. 6.
Validation of the transcriptomic data by RT-qPCR for a set of selected genes. For each gene, two different scatter plots are presented. The left panel shows the relationship between the normalized (z-score) expression levels measured by transcriptomics with microarrays (x-axis) and by RT-qPCR (y-axis). The solid line represents the null hypothesis of equal expression values (i.e., a perfect match between both quantifications). The Pearson’s correlation coefficient (r) is shown on the top of each panel. The right panel shows the correlation plots between gene expression (in red measured by microarray and in blue by RT-qPCR) and viral fitness. The solid lines represent linear models; the closer the slopes of both lines, the more similarity between microarray and RT-qPCR expression data. (A) Genes whose expression increases with viral fitness (cases from fig. 5A, red dots). (B) Genes whose expression decreases with viral fitness (cases from fig. 5B, blue dots). Bidimensional error bars represent ±1 SD. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)