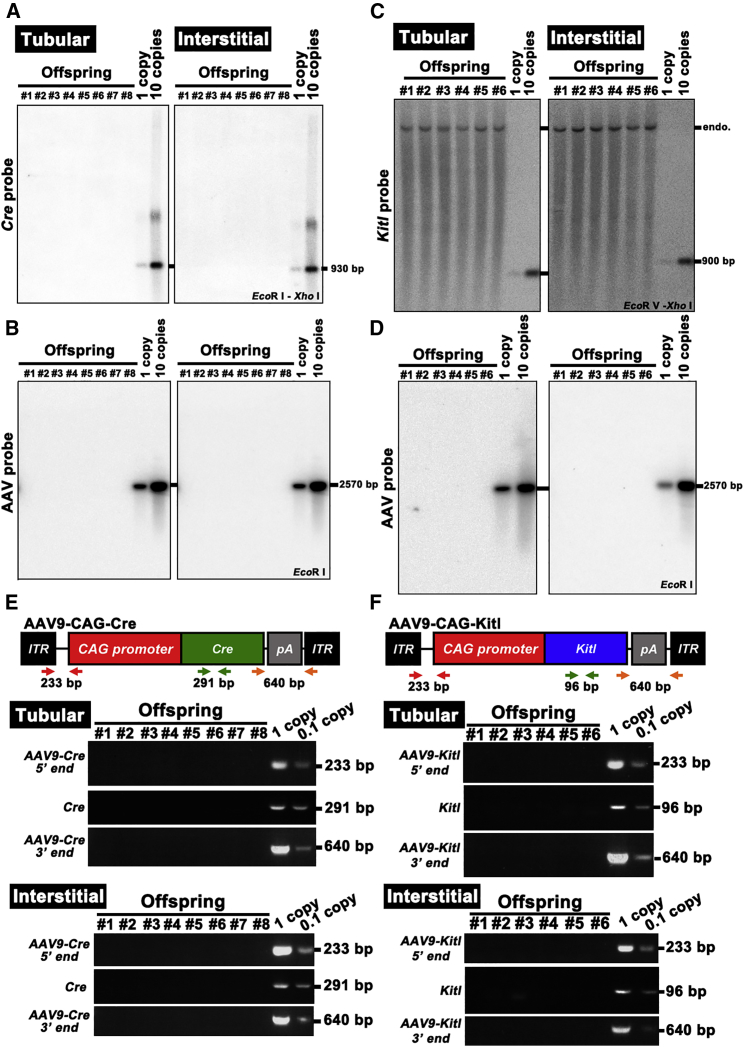
Figure 5.
Analysis of AAV9 Integration in the Offspring
(A–D) Southern blot analysis of F1 DNA samples from R26R-Eyfp (A and B) or KitlSl/KitlSl-d (C and D) mice hybridized with Cre (A), Kitl (C), or empty AAV vector (B and D). Controls represent viral DNA in amounts equivalent to 1 and ten copies of viral DNA per diploid genome. DNA was digested with the indicated restriction enzymes. Note endogenous (endo.) Kitl locus produced bands in all samples in KitlSl/KitlSl-d mouse experiment.
(E) PCR analysis of AAV9-Cre integration in the tail DNA. AAV9-specific 5′- or 3′-end-specific primers as well as Cre-specific primers were used. Samples were collected from offspring produced after tubular or interstitial injection with AAV9-Cre.
(F) PCR analysis of AAV9-Kitl integration in the tail DNA. AAV9-specific 5′- or 3′-end-specific primers as well as Kitl-specific primers were used. Samples were collected from offspring produced after tubular or interstitial injection with AAV9-Kitl. Controls containing 0.1 mg of normal mouse DNA were spiked with viral DNA representing 0.1 and 1 copies of the viral genome.
See also Table S2.