Figure 1.
Genome-Scale Methylation Dynamics during Male and Female Cell Reprogramming
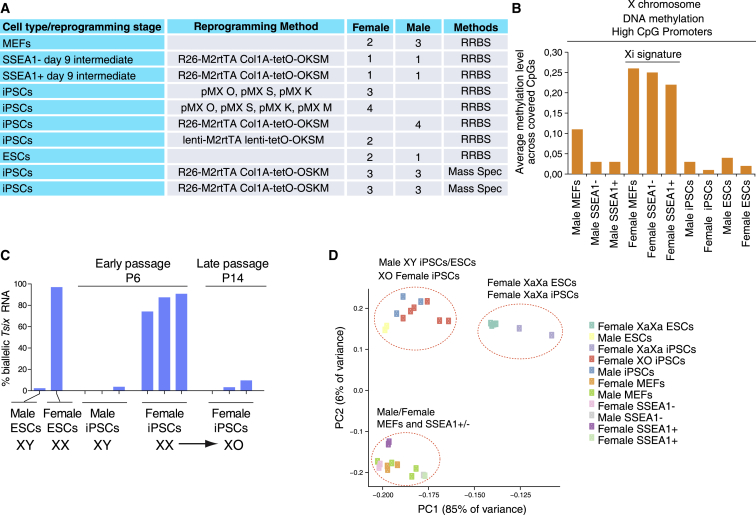
(A) Detailed information of the cell types and reprogramming methods used, and the analysis method for methylation. Further details are included in Table S1.
(B) Average methylation across listed male and female cell types and reprogramming stages at high CpG-density promoters (HCPs) on the X chromosome. The Xi signature highlights the hemi-methylation of the inactive X chromosome (Xi) present specifically in female MEFs and SSEA1+/− reprogramming stages.
(C) RNA FISH analysis of Tsix biallelic expression. The counts are presented as percentage of cells with Tsix signal (monoallelic or biallelic) that show biallelic Tsix signals. The female iPSC lines analyzed at passage 6 are the same lines as those analyzed at passage 14. The same lines at passages 5, 7, and 13 were also used in Figure 2E.
(D) PCA for RRBS-derived methylation data, considering male and female MEFs, male and female SSEA1− and SSEA1+ sorted reprogramming intermediates, male iPSCs, and female XX and XO iPSCs, as well as male and female ESCs. Each color represents a cell type and each spot represents a biological replicate (different samples). PCA was performed for the mean methylation of non-repetitive autosomal 100-bp genomic tiles with at least two CpGs covered at 5× or greater.