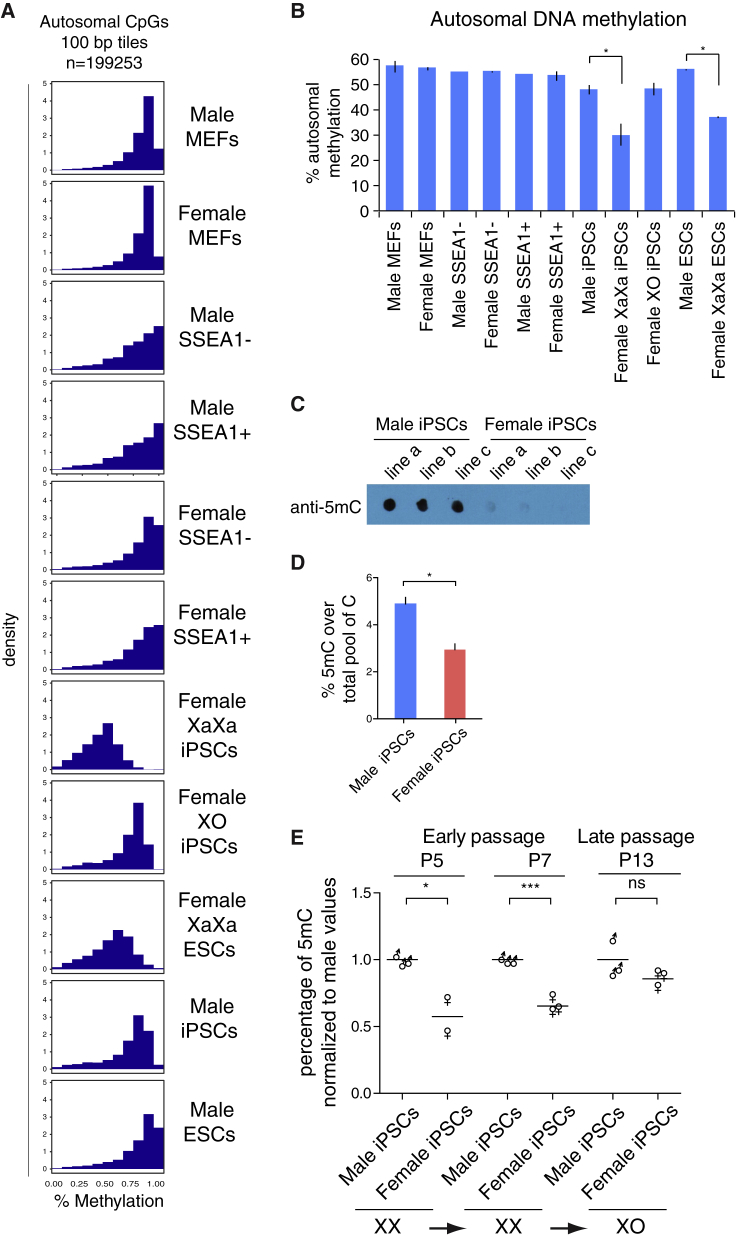
Figure 2.
Global Gain in Genome Methylation upon Loss of One of the Two X Chromosomes in iPSCs
(A) Histograms showing the distribution of methylation levels of 100-bp tiles captured by RRBS genome wide for the indicated male and female cell types and reprogramming intermediates. Only 100-bp tiles on autosomes, but not on sex chromosomes, with at least two CpGs at coverage 5× or greater were considered for this analysis. The methylation level ranges from 0 (no methylation) to 1 (100% methylation).
(B) Mean autosomal CpG methylation level over covered CpGs (1-kb tiles) across autosomes in indicated male and female cell types and reprogramming stages. Two-tailed unpaired t test, ∗p < 0.01. Error bars are SEM. The test was performed on male iPSCs and XaXa iPSCs only.
(C) Dot blot analysis of 5mC in three female and three male iPSC lines.
(D) Mass spectrometry analysis of 5mC in iPSCs. 5mC content is expressed as the percentage of 5mC in the total pool of cytosine for six male and five female iPSC lines. Data are presented as mean + SEM. Two-tailed unpaired t test, n = 2 experiments, ∗p < 0.0005.
(E) Mass spectrometry analysis of 5mC in iPSCs at different passages. 5mC content is expressed as the percentage of 5mC in the total pool of cytosine, normalized to the mean 5mC level in male samples in each time point analyzed. The average for 3 male and 2–3 female lines are shown for different passages. Two-tailed unpaired t test, n = 2 experiments, ∗p < 0.05, ∗∗∗p < 0.001; ns, not significant. The XaXa and XO state of the cell lines used was quantified for the same lines in Figure 1C.