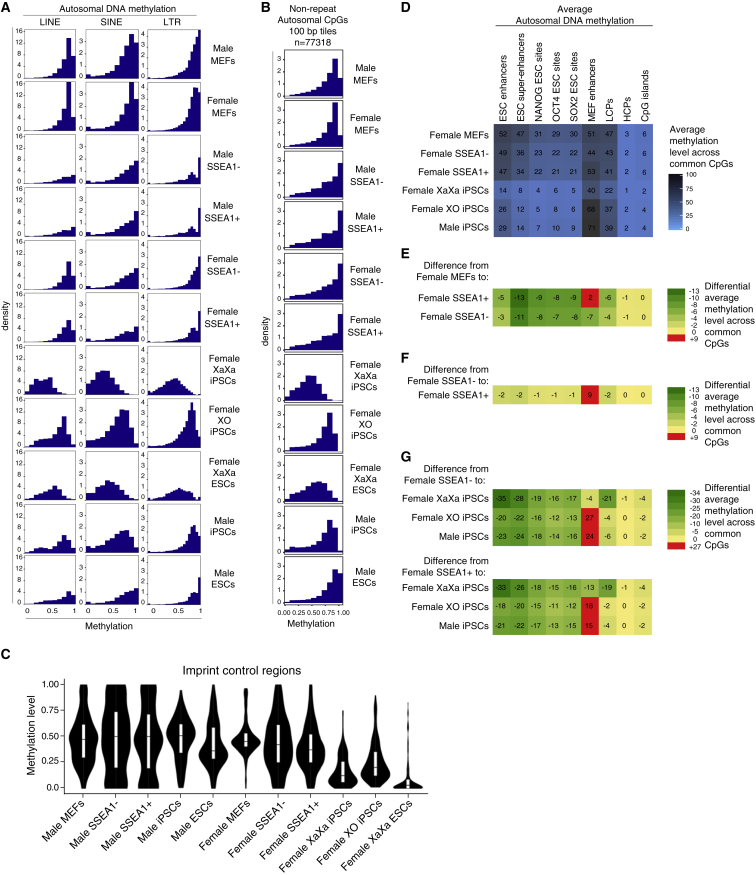
Figure 3.
Changes in Methylation at Repeat Elements, ICRs, and cis-Regulatory Sites during Reprogramming
(A) As in Figure 1C but for LINEs, SINEs, and LTRs.
(B) Histograms showing the distribution of methylation levels of non-repeat CpGs captured by RRBS genome wide for the indicated male and female cell types and reprogramming intermediates. Only 100-bp tiles on autosomes, but not on sex chromosomes, with at least two CpGs with coverage >5× across all cell types/reprogramming stages were filtered with repeat masker to exclude repeat regions. The methylation level ranges from 0 (no methylation) to 1 (100% methylation).
(C) Distributions of CpG methylation levels for ICRs, shown as violin plots.
(D) Average autosomal CpG methylation values for various cis-regulatory sites in female reprogramming and female XX and XO iPSCs as well as male iPSCs for comparison, as indicated. Coverage-weighted mean methylation was computed for each feature using CpGs with coverage 5× or greater. LCPs and HCPs refer to low and high CpG-density promoters, respectively.
(E) Heatmap of changes in average methylation for features shown in (D) for the transition from female MEFs to female SSEA1− and SSEA1+ sorted reprogramming subpopulations. The scale indicates the extent of loss or gain in average methylation. Numbers represent percent methylation.
(F) As in (E), except for the transition from female SSEA1− to the female SSEA1+ reprogramming stages.
(G) As in (E), except for the transition from female SSEA1− and SSEA1+ stages, respectively, to female XaXa iPSCs, female XO iPSCs, and male iPSCs, respectively. Note that the color scale is different from (E) and (F) but the numbers can be compared directly across (D) to (G).