Abstract
Background
Osimertinib, a third-generation epidermal growth factor receptor-tyrosine kinase inhibitor, exerts remarkable effects against EGFR T790M resistance mutation-positive non-small cell lung cancer. Identifying T790M mutation by re-biopsy is essential before prescribing osimertinib. Tissue biopsy is the golden standard for this purpose, but several factors limit its success rate. The liquid biopsy with blood, using circulating tumor DNA, has been an alternative method. However, the true biological meaning and equivalence of liquid biopsy and tumor biopsy are still under investigation. Especially, the usefulness of serum samples to detect T790M mutation is not yet been known.
Patients and methods
We prospectively evaluated the sensitivity, specificity, and parallelism of the detection of EGFR mutations in tissue re-biopsy and liquid biopsy (plasma and serum), simultaneously, from June 2016 to May 2017. EGFR mutations in tumor re-biopsy were evaluated by COBAS ver2 and PNA-LNA PCR clamp method, and those in liquid biopsy were evaluated with COBAS ver2.
Results
Fifteen patients were enrolled. In 10 patients whose EGFR mutation was detected in liquid biopsy, the original EGFR mutation (exon 19 del or L858R) was detected in all patients. Detection of EGFR mutation by COBAS ver2 and by PNA-LNA method was almost the same in tissue re-biopsy. The detection rate of T790M was lower than that of the original EGFR mutation in liquid biopsy compared to that in tissue re-biopsy. The detection of T790M in serum exhibited a higher specificity (67%) and positive predictive value (50%) than that in plasma (50% and 40%, respectively). The detection sensitivity was similar in plasma and serum.
Conclusion
Plasma, serum, and tissue genotyping can have complementary roles for detecting EGFR-T790M using COBAS ver2. Repeated tests with different samples and different methods may improve accuracy of T790M detection and will lead to the maximum benefit for the patient.
Keywords: EGFR, T790M mutation, liquid biopsy, COBAS ver2, PNA-LNA PCR clamp method, osimertinib
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. Non-small cell lung cancer (NSCLC) accounts for around 85% of all lung cancers, and majority of patients with NSCLC present with locally advanced or metastatic disease.1–3 The discovery of epidermal growth factor receptor (EGFR) mutations in NSCLC led to a new treatment paradigm. Prospective clinical trials of first- or second-generation (1st and 2nd) EGFR-tyrosine kinase inhibitors (TKIs) demonstrated remarkable response rates of approximately 70%.4–10 However, lung cancer cells inevitably acquire resistance to these inhibitors after approximately 1 year.4,8,11 The major mechanism of acquired resistance is the EGFR T790M gatekeeper mutation, which accounts for approximately half of all acquired resistance to 1st- and 2nd-generation EGFR-TKIs.12–14 Recently, third-generation EGFR-TKIs such as osimertinib have been shown to exert remarkable effects against EGFR T790M mutation-positive NSCLC. Third-generation EGFR-TKIs can bind irreversibly to the EGFR kinase by targeting the cysteine-797 residue in the ATP binding site via covalent bond formation.15 Therefore, identifying T790M mutation is very important for appropriate treatment.16–18 Re-biopsy could provide further information regarding the genetic profile of tumors and may help in identifying patients eligible for treatment.19 Tissue biopsy is the golden standard method for re-biopsy, but several factors limit its success rate such as the difficulty of accessing tumor sites and the invasive nature of the sampling methods.20–25 Given these limitations, the use of circulating tumor DNA (ctDNA), commonly named “liquid biopsy”, has emerged as a new promising and less invasive tool to detect EGFR mutations in patients with NSCLC and can provide similar molecular information as those obtained by invasive tumor biopsies.26
In June 2016, the US Food and Drug Administration approved COBAS ver2 using plasma specimens as a companion diagnostic test for the detection of exon 19 deletions or exon 21 substitution mutations in the EGFR gene, and liquid biopsy for the detection of T790M has been acknowledged as one of the re-biopsy methods in Japan since December 2016. Conversely, peptide nucleic acid-locked nucleic acid polymerase chain reaction (PNA-LNA PCR) clamp method is a laboratory-developed test to detect EGFR mutations, including T790M. Although its sensitivity is relatively high, no study has clarified which method, PNA-LNA PCR method or COBAS ver2 method, has a higher detection rate of T790M mutation in tissue and blood.
Although the usefulness of liquid biopsy for detecting EGFR mutations has been recognized, the true biological meaning and equivalence of the detection methods are still under investigation. Especially, the usefulness of serum samples to detect T790M mutation has not yet been clarified.
In this study, we prospectively evaluated the sensitivity, specificity, and parallelism of the detection of EGFR mutations in liquid biopsy (plasma and serum samples) and tumor re-biopsy in patients with EGFR-TKI-resistant NSCLC using COBAS ver2 and PNA-LNA PCR clamp method. We aimed to identify clinical characteristics that predict the high T790M-positive rate in liquid biopsy. We also evaluated the usefulness and significance of serum testing in liquid biopsy to detect T790M mutation.
Patients and methods
Patients
From June 2016 to May 2017, we enrolled patients with advanced or recurrent EGFR-mutant NSCLC who underwent re-biopsy (2nd biopsy) based on physician’s decision in the cases of acquired resistance to 1st- or 2nd-generation EGFR-TKI treatment. All tissue samples were obtained from sites assessed as disease progression by imaging. All patients were enrolled in the Keio University Hospital. Acquired resistance was defined according to Jackman’s criteria.27 Written informed consent was obtained from all patients. We also obtained consent from patients regarding the publication of images. This study was approved by the Ethical Review Board Committee of Keio University. Research was conducted in accordance with the 1964 Declaration of Helsinki, as revised in 2013.
Tissue re-biopsy and liquid biopsy
We prospectively evaluated the sensitivity, specificity, and parallelism of the detection of EGFR mutations in tissue re-biopsy and liquid biopsy (plasma and serum) simultaneously. EGFR mutations in tumor re-biopsy were evaluated with COBAS ver2 and PNA-LNA PCR clamp method and those in liquid biopsy were evaluated with COBAS ver2 using plasma and serum. EGFR mutation detection in liquid biopsy was carried out by BML Inc. (Tokyo, Japan).
The amount of blood needed for plasma analysis was approximately 12 mL with EDTA as an anticoagulant. Plasma was separated from the blood cells at 4°C via low-speed centrifugation (1,200× g for 15 min) within 4 hours after blood draw. The pre-analytical (sample collection and processing) protocols for serum samples are simpler than those for plasma. Approximately 9 mL of blood was harvested in a normal test tube and the serum was separated from blood cells at room temperature using the same centrifugation method. These liquid samples were stored at −80°C until DNA extraction.
Clinical characteristics
The following clinical characteristics were included in our analyses: age, sex, smoking status, the presence or absence of bevacizumab before 2nd biopsy, treatment duration with EGFR-TKIs (gefitinib, erlotinib, and afatinib), interval between diagnosis and 2nd biopsy, and the number of metastases before re-biopsy. To determine the number of metastatic sites, we counted “only one rib metastasis” as “one bone metastasis” and “one rib, one iliac, and one skull metastasis” as “three bone metastases.”
Efficacy of osimertinib
Responsiveness to osimertinib was evaluated according to the Response Evaluation Criteria in Solid Tumors (version 1.1).28
Statistical analysis
All categorical variables were analyzed by Fisher’s exact test or Student’s t-test, as appropriate. All p-values <0.05 were considered statistically significant. Statistical analyses were conducted by using JMP software (version 13; SAS Institute, Cary, NC, USA). Data were visualized by using Easy R (Jichi Medical University, Saitama, Japan), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria).29
Results
Patient characteristics
Fifteen patients with NSCLC harboring EGFR mutations were enrolled in this study. They underwent re-biopsy after acquiring resistance to EGFR-TKIs. Patient characteristics are shown in Table 1. The median age was 62 years, and majority of the original EGFR mutations found were exon 19 deletion (53%) and exon 21 L858R (33%).
Table 1.
Patient characteristics (n=15)
| Age, year, median (range) | 62 (44–84) |
|---|---|
| Sex, n (%) | |
| Male | 5 (33) |
| Female | 10 (67) |
| Original mutation, n (%) | |
| Exon 19 deletion | 8 (53) |
| L858R | 5 (33) |
| G719C | 1 (7) |
| G719S | 1 (7) |
| Smoking status, n (%) | |
| Never | 8 (53) |
| Former | 7 (47) |
| EGFR-TKIs, n | |
| Erlotinib | 14a |
| Gefitinib | 4 |
| Afatinib | 4 |
| Bevacizumab as prior therapy, n (%) | |
| Yes | 9 (60) |
| Duration of erlotinib treatment, m, median (range) | 10 (0–29) |
| Duration of all EGFR-TKIs treatment, m, median (range) | 23 (3–59) |
| Interval between diagnosis and re-biopsy, m, median (range) | 35 (10–140) |
| Number of metastases, n, median (range) | 3 (0–9) |
Notes:
Seven patients were treated with double EGFR-TKIs. Eleven patients were treated with erlotinib as the initial line of EGFR-TKIs.
Abbreviations: EGFR-TKIs, epidermal growth factor receptor-tyrosine kinase inhibitors; m, months.
Fourteen patients were treated with erlotinib, four patients with gefitinib, and four patients with afatinib. Seven patients were treated with double EGFR-TKIs. Eleven patients were treated with erlotinib as the initial EGFR-TKI. Nine patients were treated with bevacizumab sometime before re-biopsy.
Ten patients presented positive results for EGFR mutations (exon 19 deletion, L858R, or T790M) in the liquid biopsy (plasma or serum) (Figure 1). Of these patients, only five had a positive result for T790M with the liquid biopsy. Eight patients had a positive result for T790M with tissue re-biopsy.
Figure 1.
Patient flow diagram.
Note: L(−) denotes no mutation detected in the liquid biopsy for both original mutations and T790M.
Abbreviations: T, tissue; L, liquid biopsy.
Differences in the methodology between the first tumor tissue biopsy and re-biopsy
There were differences in the methods used between the first biopsy (ie, at the time of first diagnosis) and re-biopsy (Table S1). CT-guided needle biopsy was used more frequently in re-biopsy than for the first biopsy.
Assay characteristics
In 10 patients whose EGFR mutation was detected in liquid biopsy, the original EGFR mutation (exon 19 deletion or L858R) was detected in all patients: both with plasma and serum. COBAS ver2 and PNA-LNA PCR clamp method exhibited 100% sensitivity, specificity, and positive predictive value for the detection of EGFR exon 19 deletion (7/7) and L858R (3/3) when compared with tumor genotyping of the re-biopsy tissue. The results of EGFR mutation detection in tissue re-biopsy using COBAS ver2 and PNA-LNA PCR clamp method were consistent, except for one case presenting a positive result with PNA-LNA PCR clamp method, but not with COBAS ver2 (Figure 2).
Figure 2.
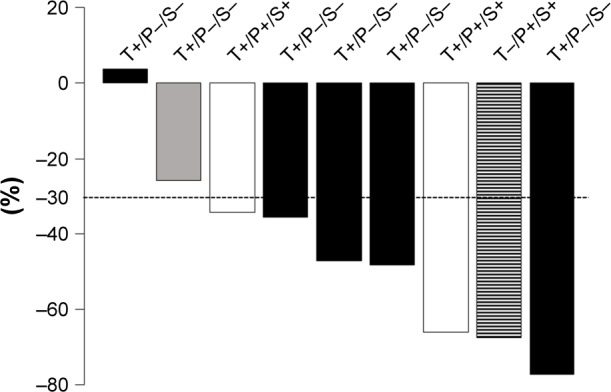
Waterfall plot of the percentage change in target lesion size at the initial response evaluation.
Notes: The patients positive for T790M mutation in tissue re-biopsy by COBAS ver2/PNA-LNA PCR clamp method and negative in liquid biopsy (plasma/serum) are represented in black; the patients positive for T790M both in tissue re-biopsy and liquid biopsy (plasma/serum) are represented in white; the patients positive for T790M in tissue re-biopsy by PNA-LNA PCR clamp method but negative by COBAS ver2 and in liquid biopsy (plasma/serum) are represented in gray; and the patients positive for T790M in liquid biopsy (plasma/serum) and negative in tissue re-biopsy are represented by the hatched bar.
Abbreviations: P, plasma; PNA-LNA PCR, peptide nucleic acid-locked nucleic acid polymerase chain reaction; S, serum; T, tissue.
By contrast, the detection rate of T790M in liquid biopsy (50%) was lower than that of major EGFR mutation (exon 19 del or L858R, 100%) (Table S2).
The detection of T790M mutation in serum exhibited a higher specificity (67% [4/6]) and positive predictive value (50% [2/4]) than that in plasma (50% [3/6], 40% [2/5], respectively). The sensitivity for the detection of T790M was similar when using plasma and serum (50% [2/4]).
Clinical factors related to T790M prevalence in liquid biopsy and tissue re-biopsy
When considering the results of the liquid biopsy, the number of metastases was significantly higher in the T790M-positive group than in the T790M-negative group in both plasma and serum (Table 2). By contrast, in tissue re-biopsy, longer duration of erlotinib treatment, but not the total duration of EGFR-TKI treatment or the number of metastases, was significantly related to T790M-positivity (Table 3).
Table 2.
Univariate analyses of patient characteristics and T790M prevalence (plasma and serum)
| Patient characteristics | T790M+ (plasma)
|
T790M− (plasma)
|
p-value | T790M+ (serum)
|
T790M− (serum)
|
p-value | |
|---|---|---|---|---|---|---|---|
| n=10 | n=5 | n=5 | n=4 | n=6 | |||
| Age, year | >65 | 2 | 3 | 1 | 2 | 3 | 1 |
| Sex, n | Male | 0 | 2 | 0.44 | 0 | 2 | 0.47 |
| Smoking status, n | Yes | 0 | 3 | 0.17 | 0 | 3 | 0.2 |
| Bevacizumab as prior therapy, n | Yes | 4 | 2 | 0.53 | 4 | 2 | 0.077 |
| Duration of erlotinib treatment, m | 7.6±3.9 | 11±3.9 | 0.56 | 8.5±4.5 | 9.8±3.7 | 0.82 | |
| Duration of all EGFR-TKIs treatment, m | 13.8±7.7 | 24.1±5.6 | 0.38 | 16.3±9.0 | 20.1±7.4 | 0.71 | |
| Interval between diagnosis and re-biopsy, m | 31.2±16.2 | 59.0±16.2 | 0.26 | 34.5±19.1 | 52.2±15.6 | 0.49 | |
| Number of metastases, n | 6.2±0.8 | 2.0±0.8 | 0.006 | 6.5±1.0 | 2.5±0.8 | 0.013 |
Note: Data are shown as mean ± SD.
Abbreviations: EGFR-TKIs, epidermal growth factor receptor-tyrosine kinase inhibitors; m, months.
Table 3.
Univariate analyses of patient characteristics and T790M prevalence in tissue biopsy
| Patient characteristics | T790M+ (tissue)
|
T790M− (tissue)
|
p-value | |
|---|---|---|---|---|
| n=9 | n=6 | |||
| Age, n | >65 year | 4 | 3 | 1 |
| Sex, n | Male | 3 | 2 | 1 |
| Smoking status, n | Yes | 4 | 4 | 1 |
| Bevacizumab as prior therapy, n | Yes | 6 | 3 | 0.31 |
| Duration of erlotinib treatment, m | 16±2.2 | 6.3±2.4 | 0.01 | |
| Duration of all EGFR-TKIs treatment, m | 19.9±5.6 | 22.3±5.6 | 0.77 | |
| Interval between diagnosis and re-biopsy, m | 52.3±12.8 | 40.3±13.7 | 0.53 | |
| Number of metastases, n | 19.9±5.3 | 18.9±5.6 | 0.9 |
Note: Data are shown as mean ± SD.
Abbreviations: EGFR-TKIs, epidermal growth factor receptor-tyrosine kinase inhibitors; m, months.
Efficacy of osimertinib
Nine patients were treated with osimertinib. The response rate was 77.8% (7/9) and disease control rate was 100% (9/9) (Figure 2). A response was observed in patients with T790M mutation detected in both tissue and liquid biopsy, only in tissue, and only in liquid biopsy. One patient was positive for T790M in liquid biopsy (both plasma and serum) and negative in tissue re-biopsy (transbronchial lung biopsy). After 2 months of gefitinib treatment, another re-biopsy of liver metastasis was performed and was positive for T790M. Afterward, the patient was treated with osimertinib and presented a response in both the lung and liver metastases (Figure 3).
Figure 3.
A representative patient with osimertinib treatment.
Notes: Chest x ray (A) prior to and (B) after treatment with osimertinib for 2 months. Abdomen computed tomography (C) prior to and (D) after treatment with osimertinib. This patient was T790M negative with tissue re-biopsy (lung) and T790M positive with liquid biopsy. Re-biopsy of liver metastasis (white arrows) revealed T790M positivity 2 months after the first lung re-biopsy. Osimertinib treatment resulted in tumor shrinkage both in the lung and liver.
Discussion
In the Phase III LUX-Lung 3/6 trials in patients with EGFR mutation-positive lung adenocarcinoma, EGFR mutation detection rates in ctDNA were 28.6% (serum) and 60.5% (plasma) by using RT-PCR (scorpion-ARMS) method.30 Previous studies suggested that the plasma may be a more reliable source than the serum for ctDNA owing to higher background levels of non-tumor wild-type DNA found in serum. Specifically, serum yielded more cell free DNA than plasma, but the absolute quantity of mutation was higher in plasma, which may explain the differences in detection.31–33 These reports suggested that the sensitivity of the detection of EGFR mutation in the serum is lower than that in the plasma when tumor re-biopsy is used as a reference. We prospectively performed the current study to elucidate the difference in the usefulness of plasma and serum. It is known that the detection of T790M mutation after acquired resistance is always accompanied by the original EGFR mutation.34 We detected high concordance in terms of the sensitivity of EGFR mutation detection between plasma and serum samples using COBAS ver2, although the detection rate of T790M was lower than that of exon 19 deletion or L858R. Our study suggests that the detection of EGFR mutations in serum samples using COBAS ver2 might be as reliable as that in plasma samples.
Tissue biopsies remain the gold standard for the detection of T790M mutation, because the AURA study revealed that the efficacy of osimertinib for patients detected as T790M positive in plasma and T790M negative in the re-biopsy tissue was lower than that for patients detected as T790M positive in the tissue. Current clinical practice indicates that performing re-biopsy at the site of relapse is necessary to predict the efficacy of osimertinib because of the heterogeneous presence of T790M in the tumor tissue.35 Indeed, in our study, one patient was diagnosed as presenting only an EGFR exon 19 deletion mutation by re-biopsy with trans-bronchial lung biopsy. However, the liquid biopsies (both plasma and serum) were positive for T790M. The patient underwent an additional re-biopsy after 2 months of targeting liver metastasis because her major relapse site was the liver. The result indicated that the patient was T790M positive. She had a remarkable response to osimertinib (Figure 3).
T790M detection at the sites of relapsed tumor tissue is required to evaluate the effectiveness of osimertinib against the relapsed tumor. Using plasma samples, we may detect a little T790M clone leakage of non-relapse tumor tissue. If this is the case, we cannot predict the efficacy of osimertinib in treating relapsed tumor sites in these patients. Our data of T790M detection in liquid biopsy revealed that the specificity and positive predictive value of T790M detection in the serum were higher than those in the plasma. Thus, if the serum genotype is positive for T790M, it is likely that the patient harbored T790M mutation in the tissue, which is highly suspected to be a relapsed tumor site. These data suggest that serum samples as liquid biopsy may not detect a little T790M clone leakage from non-relapse sites. Serum samples were more reliable in the detection of T790M mutation at the relapse tumor tissue sites than plasma samples. We also suggest serum samples to be more useful than plasma samples in the detection of EGFR T790M mutation for determining the effectiveness of osimertinib against relapse tumor sites. If only liquid biopsy is conducted for the detection of T790M because tissue re-biopsy cannot be performed due to difficulties in accessing the tumor sites or because tumor tissue re-biopsy was negative for T790M, it might be useful to rely on the result of both serum and plasma samples. We suggest that the combined results of mutation detection using serum and plasma could provide insights into the efficacy of osimertinib treatment.
The major limitation of our study was the small sample size. Further enrollment of cases is necessary. However, we believe that the heterogeneous patient population in our study may be very important and represent population variability, resembling other studies’ populations. First, the number of metastases was significantly higher in the T790M-positive group than in the T790M negative group (Table 2). Thress et al reported that T790M mutation was more readily detected in the plasma of patients with extra-thoracic metastatic disease than in that from patients with disease confined to the thoracic cavity (as determined by droplet digital PCR assay).36 This suggests that tumor bulk and metastatic status may impact on the presence EGFR mutation in the plasma and should be further validated in clinical settings.37 We also found that the duration of erlotinib treatment was significantly longer in the T790M-positive group than in the T790M-negative group. Several reports analyzed the relationship between T790M induction and exposure to EGFR-TKIs in preclinical and clinical studies.38–40 We do not have an explanation as to why only the duration of erlotinib treatment, but not the total duration of the TKI treatment, is related to T790M positivity. One possibility may be that the total duration includes “beyond PD” treatment, ie, EGFR-TKI treatment after progression, and may not relate to the induction of T790M. Further studies are needed to confirm this possibility.
Finally, we found a case with discordant results of T790M detection in tissue re-biopsy using COBAS ver2 and PNA-LNA PCR clamp method. Previous studies reported that the detection rate of EGFR mutation is higher with the PNA-LNA PCR clamp method than with COBAS ver2.41,42 Although the companion diagnostic is COBAS ver2, one might consider using a different method like PNA-LNA PCR clamp method to confirm the T790M negative results by COBAS ver2, to detect T790M, in order to not directly rule out the possibility of osimertinib in T790M-negative cases by COBAS ver2.
Conclusion
Our data suggest that plasma, serum, and tissue genotyping can have complementary roles for T790M testing by using COBAS ver2 and the PNA-LNA PCR clamp method. Especially, the result of both serum and plasma samples for T790M detection could be more useful to predict the efficacy of osimertinib than only plasma samples when tissue genotyping is negative for T790M or tissue genotyping is unknown.
Supplementary materials
Table S1.
Comparison between the first biopsy and re-biopsy
| Site of biopsy | First biopsy n (%) | Re-biopsy n (%) |
|---|---|---|
| Biopsy | ||
| Lung | ||
| Bronchoscopy | 8 (53) | 3 (20) |
| CT-NB | 4 (27) | 8 (53) |
| Lymph node | ||
| Bronchoscopy | 1 (7) | 0 |
| CT-NB | 0 | 2 (13) |
| Bone | ||
| CT-NB | 1 (7) | 0 |
| Liver | ||
| CT-NB | 0 | 1 (7) |
| Surgery | ||
| CNS | 1 (7) | 0 |
| Fluid | ||
| Pleural effusion | 0 | 1 (7) |
| Biopsy sites | ||
| Primary | 13 (87) | 8 (53) |
| Metastasis | 2 (13) | 7 (47) |
Abbreviations: CNS, central nervous system; CT-NB, computed tomography-guided needle biopsy.
Table S2.
Plasma and serum assay sensitivity, specificity, and positive predictive value
| Mutation | Sensitivity, % | True positive, n | False negative, n | Specificity, % | True negative, n | False positive, n | Positive predictive value, % |
|---|---|---|---|---|---|---|---|
| Exon 19 del | |||||||
| Plasma | 100 | 7 | 0 | 100 | 3 | 0 | 100 |
| Serum | 100 | 7 | 0 | 100 | 3 | 0 | 100 |
| L858R | |||||||
| Plasma | 100 | 3 | 0 | 100 | 7 | 0 | 100 |
| Serum | 100 | 3 | 0 | 100 | 7 | 0 | 100 |
| T790M | |||||||
| Plasma | 50 | 2 | 2 | 50 | 3 | 3 | 40 |
| Serum | 50 | 2 | 2 | 67 | 4 | 2 | 50 |
Acknowledgments
We would like to thank Ms Kimiko Shibuya for sample preparation and BML Inc., Tokyo, Japan, for sample analysis. KN was funded by the Grant-in-Aid for Scientific Research, Japan Society for Promotion of Science.
Footnotes
Author contributions
KK participated in the acquisition of data, analysis and interpretation of data, and drafting the manuscript and revising it. KN participated in the conception and design, acquisition of data, analysis and interpretation of data, and drafting the manuscript and revising it. TM, KM, HH, HY, IK, KS, and TB participated in the interpretation of data and drafting the manuscript and revising it. All the authors read and approved the final manuscript and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359(13):1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuda T, Marugame T, Kamo K, Katanoda K, Ajiki W, Sobue T, Japan Cancer Surveillance Research Group Cancer incidence and incidence rates in Japan in 2002: based on data from 11 population-based cancer registries. Jpn J Clin Oncol. 2008;38(9):641–648. doi: 10.1093/jjco/hyn074. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 4.Maemondo M, Inoue A, Kobayashi K, et al. North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T, Morita S, Yatabe Y, et al. West Japan Oncology Group Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised Phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 6.Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a Phase III, randomized, open-label, first-line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non-small-cell lung cancer in Asia (IPASS) J Clin Oncol. 2011;29(21):2866–2874. doi: 10.1200/JCO.2010.33.4235. [DOI] [PubMed] [Google Scholar]
- 7.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, Phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 8.Rosell R, Carcereny E, Gervais R, et al. Spanish Lung Cancer Group in Collaboration with Groupe Français de Pneumo-Cancérologie and Associazione Italiana Oncologia Toracica Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised Phase 3 trial. Lancet Oncol. 2012;13(3):239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 9.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 10.Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised Phase 3 trial. Lancet Oncol. 2014;15(2):213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 11.Miller VA, Hirsh V, Cadranel J, et al. Afatinib versus placebo for patients with advanced, metastatic non-small-cell lung cancer after failure of erlotinib, gefitinib, or both, and one or two lines of chemotherapy (LUX-Lung 1): a Phase 2b/3 randomised trial. Lancet Oncol. 2012;13(5):528–538. doi: 10.1016/S1470-2045(12)70087-6. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19(8):2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russo A, Franchina T, Ricciardi GRR, et al. Third generation EGFR TKIs in EGFR-mutated NSCLC: where are we now and where are we going. Crit Rev Oncol Hematol. 2017;117:38–47. doi: 10.1016/j.critrevonc.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med. 2015;372(18):1689–1699. doi: 10.1056/NEJMoa1411817. [DOI] [PubMed] [Google Scholar]
- 17.Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2015;372(18):1700–1709. doi: 10.1056/NEJMoa1413654. [DOI] [PubMed] [Google Scholar]
- 18.Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4(9):1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suda K, Murakami I, Sakai K, et al. Small cell lung cancer transformation and T790M mutation: complimentary roles in acquired resistance to kinase inhibitors in lung cancer. Sci Rep. 2015;5:14447. doi: 10.1038/srep14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirita K, Izumo T, Matsumoto Y, Hiraishi Y, Tsuchida T. Bronchoscopic re-biopsy for mutational analysis of non-small cell lung cancer. Lung. 2016;194(3):371–378. doi: 10.1007/s00408-016-9864-5. [DOI] [PubMed] [Google Scholar]
- 21.Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re-biopsy in advanced non small-cell lung cancer: a prospective multi-center study in a real-world setting (GFPC study 12-01) Lung Cancer. 2014;86(2):170–173. doi: 10.1016/j.lungcan.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa T, Sawa T, Futamura Y, et al. Feasibility of rebiopsy in non-small cell lung cancer treated with epidermal growth factor receptor-tyrosine kinase inhibitors. Intern Med. 2015;54(16):1977–1980. doi: 10.2169/internalmedicine.54.4394. [DOI] [PubMed] [Google Scholar]
- 23.Kawamura T, Kenmotsu H, Taira T, et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci. 2016;107(7):1001–1005. doi: 10.1111/cas.12963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol. 2013;31(1):17–22. doi: 10.1200/JCO.2012.43.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) Lancet. 2016;387(10026):1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 26.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32(6):579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackman D, Pao W, Riely GJ, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28(2):357–360. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92(3):205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 29.Kanda Y. Investigation of the freely available easy-to-use software “EZR” for medical statistics. Bone Marrow Transplant. 2013;48(3):452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu YL, Sequist LV, Hu CP, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer. 2017;116(2):175–185. doi: 10.1038/bjc.2016.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinman CR. Free DNA in serum and plasma from normal adults. J Clin Invest. 1975;56(2):512–515. doi: 10.1172/JCI108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Board RE, Williams VS, Knight L, et al. Isolation and extraction of circulating tumor DNA from patients with small cell lung cancer. Ann N Y Acad Sci. 2008;1137:98–107. doi: 10.1196/annals.1448.020. [DOI] [PubMed] [Google Scholar]
- 33.Mayo-de-Las-Casas C, Jordana-Ariza N, Garzón-Ibañez M, et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann Oncol. 2017;28(9):2248–2255. doi: 10.1093/annonc/mdx288. [DOI] [PubMed] [Google Scholar]
- 34.Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer. 2014;120(24):3896–3901. doi: 10.1002/cncr.28964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zarogoulidis P, Gaga M, Huang H, Darwiche K, Rapti A, Hohenforst-Schmidt W. Tissue is the issue and tissue competition. Re-biopsy for mutation T790: where and why? Clin Transl Med. 2017;6(1):6. doi: 10.1186/s40169-017-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–515. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 37.Remon J, Menis J, Hasan B, et al. The APPLE trial: feasibility and activity of AZD9291 (osimertinib) treatment on positive plasma T790M in EGFR-mutant NSCLC patients. EORTC 1613. Clin Lung Cancer. 2017;18(5):583–588. doi: 10.1016/j.cllc.2017.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Ogino A, Kitao H, Hirano S, et al. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67(16):7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- 39.Matsuo N, Azuma K, Sakai K, et al. Association of EGFR exon 19 deletion and EGFR-TKI treatment duration with frequency of T790M mutation in EGFR-mutant lung cancer patients. Sci Rep. 2016;6:36458. doi: 10.1038/srep36458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA analysis for detection of the EGFR T790M mutation in patients with advanced non-small cell lung cancer. J Thorac Oncol. 2017;12(7):1061–1070. doi: 10.1016/j.jtho.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Lopez-Rios F, Angulo B, Gomez B, et al. Comparison of molecular testing methods for the detection of EGFR mutations in formalin-fixed paraffin-embedded tissue specimens of non-small cell lung cancer. J Clin Pathol. 2013;66(5):381–385. doi: 10.1136/jclinpath-2012-201240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costa C, Molina MA, Drozdowskyj A, et al. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized Phase III EURTAC trial. Clin Cancer Res. 2014;20(7):2001–2010. doi: 10.1158/1078-0432.CCR-13-2233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Comparison between the first biopsy and re-biopsy
| Site of biopsy | First biopsy n (%) | Re-biopsy n (%) |
|---|---|---|
| Biopsy | ||
| Lung | ||
| Bronchoscopy | 8 (53) | 3 (20) |
| CT-NB | 4 (27) | 8 (53) |
| Lymph node | ||
| Bronchoscopy | 1 (7) | 0 |
| CT-NB | 0 | 2 (13) |
| Bone | ||
| CT-NB | 1 (7) | 0 |
| Liver | ||
| CT-NB | 0 | 1 (7) |
| Surgery | ||
| CNS | 1 (7) | 0 |
| Fluid | ||
| Pleural effusion | 0 | 1 (7) |
| Biopsy sites | ||
| Primary | 13 (87) | 8 (53) |
| Metastasis | 2 (13) | 7 (47) |
Abbreviations: CNS, central nervous system; CT-NB, computed tomography-guided needle biopsy.
Table S2.
Plasma and serum assay sensitivity, specificity, and positive predictive value
| Mutation | Sensitivity, % | True positive, n | False negative, n | Specificity, % | True negative, n | False positive, n | Positive predictive value, % |
|---|---|---|---|---|---|---|---|
| Exon 19 del | |||||||
| Plasma | 100 | 7 | 0 | 100 | 3 | 0 | 100 |
| Serum | 100 | 7 | 0 | 100 | 3 | 0 | 100 |
| L858R | |||||||
| Plasma | 100 | 3 | 0 | 100 | 7 | 0 | 100 |
| Serum | 100 | 3 | 0 | 100 | 7 | 0 | 100 |
| T790M | |||||||
| Plasma | 50 | 2 | 2 | 50 | 3 | 3 | 40 |
| Serum | 50 | 2 | 2 | 67 | 4 | 2 | 50 |




