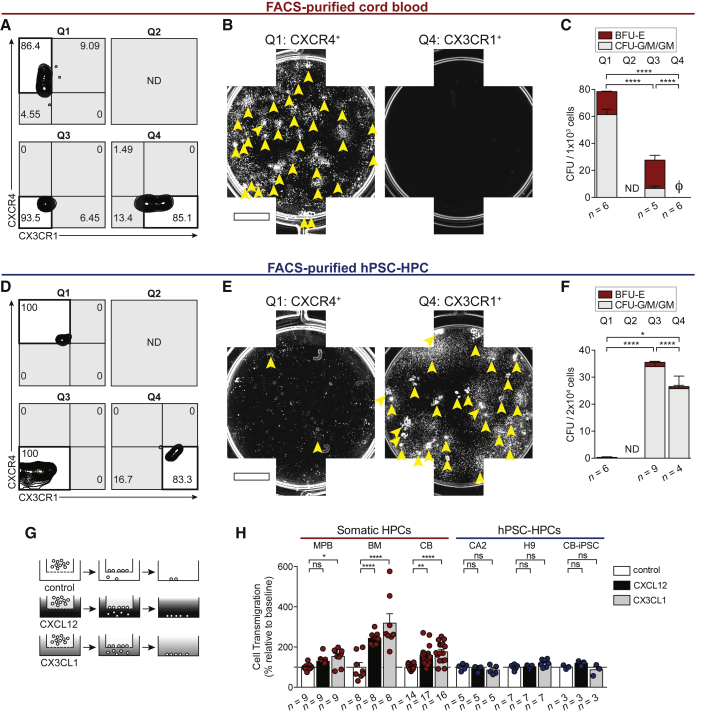
Figure 4.
Differential Segregation of Progenitors by CXCR4 and CX3CR1
(A) Sort purities from CB cells FACS-purified by CXCR4 and CX3CR1 expression. Quadrant (Q) 2 was not detected (ND).
(B) Composite well images of Q1- and Q4-sorted CB. 5 × 102 Q1 and 1 × 103 Q4 sorted cells were seeded per well. Arrowheads indicate individual CFU. Scale bar represents 5 mm.
(C) CFU frequency of FACS-purified CB based on chemokine receptor expression. Data points represent n independently assayed wells (precise n values indicated in the figure). Ø is zero. Q2 was ND. Data are represented as means ± SEM.
(D) Sort purities from hPSC-derived hematopoietic cells FACS-purified by CXCR4 and CX3CR1 expression. Q2 was ND.
(E) Composite well images of Q1- and Q4-sorted hPSC-derived hematopoietic cells; 2 × 104 sorted cells were seeded per well. Arrowheads indicate individual CFU. Scale bar represents 5 mm.
(F) CFU frequency of FACS-purified hPSC-derived hematopoietic cells based on chemokine receptor expression. Data points represent n independently assayed wells (precise n values indicated in the figure). Q2 was ND. Data are represented as means ± SEM.
(G) Transwell assay was conducted with 200 ng/mL CXCL12, or 200 ng/mL CX3CL1, or control (0.001% BSA).
(H) Transwell migration was quantified by flow cytometry. Data points represent n independently assayed wells (precise n values indicated in the figure), pooled from four independently performed experiments. CB used as positive control in every hPSC-HPC experiment. Two-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Data are represented as means ± SEM.