Figure 5.
Overcoming Resistance to CXCR4 Expression Enables CXCL12-Dependent Signaling
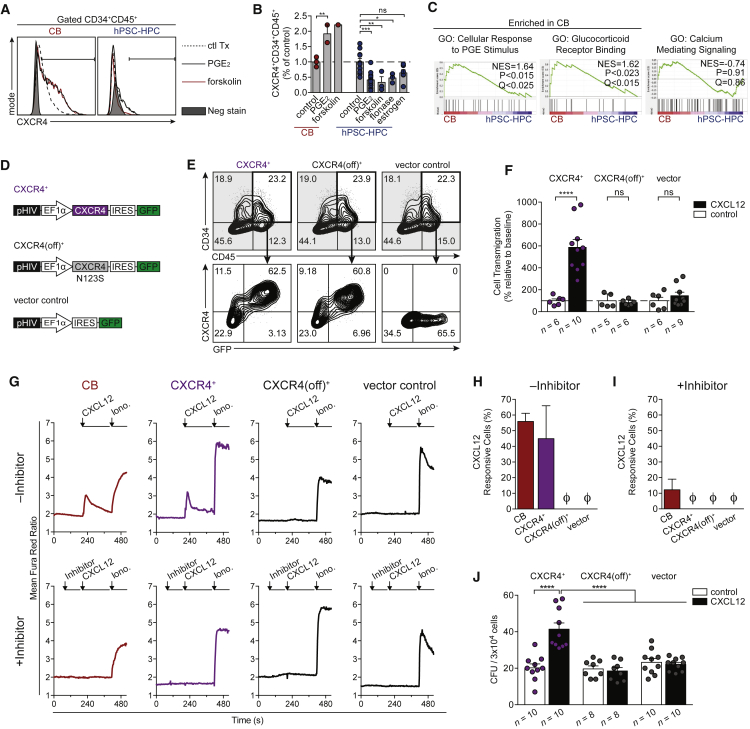
(A) Flow cytometric of CXCR4 within CD34+CD45+ CB or hPSC-HPCs treated with indicated compounds; see also Supplemental Experimental Procedures.
(B) A summary of total CXCR4+CD34+CD45+ cells from CB and hPSC-HPCs, relative to control (0.01% DMSO or BSA). Data points represent n independently assayed wells (precise n values indicated in Supplemental Experimental Procedures), pooled from three independently performed experiments. Data are represented as means ± SEM. Two-way ANOVA, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) GSEA analysis of pathways that regulate CXCR4. Samples: CB and H9-HPC; CD34+CD45+CD38–, shown in Figure 3A. GO, Gene Ontology; NES, normalized enrichment score.
(D) Human CXCR4 was cloned into pHIV-EGFP, which has an internal ribosome entry site (IRES) translation link to GFP (CXCR4+). Site-directed mutagenesis of CXCR4 (N123K) was termed CXCR4(off)+. Vector control, expressing GFP, was used in parallel for all experiments.
(E) Flow cytometry at 48 hr post transduction on EB day 16, showing comparable hPSC-HPC frequency (CD34+CD45+), and robust CXCR4+GFP+ co-expression.
(F) Transwell assay was conducted with 200 ng/mL CXCL12 or control (0.001% BSA), and quantified by flow cytometry at 48 hr post transduction on EB day 16. Data points represent n independently assayed wells (precise n values indicated in the figure), pooled from three independently performed experiments. Two-way ANOVA, ∗∗∗∗p < 0.0001. Data are represented as means ± SEM.
(G) Calcium flux transients were monitored in response to CXCL12 (200 ng/mL) in the absence (top row) or presence (bottom row) of CXCR4 inhibitor (AMD3100, 10 μM) at 48 hr post transduction on EB day 16. Ionomycin (10 μM) treatment was used as a positive control to identify live cells.
(H and I) The frequency of cells that responded to CXCL12 treatment (transient >150% of baseline) was quantified in the absence (H) or presence (I) of CXCR4 inhibitor, AMD3100. Data points represent n = 4 independently assayed wells per sample, pooled from two independently performed experiments. Ø is zero. Data are represented as means ± SEM.
(J) Total CFU counts from transduced hPSC-HPCs seeded into MethoCult ±150 ng/mL CXCL12 at 48 hr post transduction on EB day 16. Data points represent n independently assayed wells (precise n values indicated in the figure), pooled from three independently performed experiments. Two-way ANOVA, ∗∗∗∗p < 0.0001. Data are represented as means ± SEM.