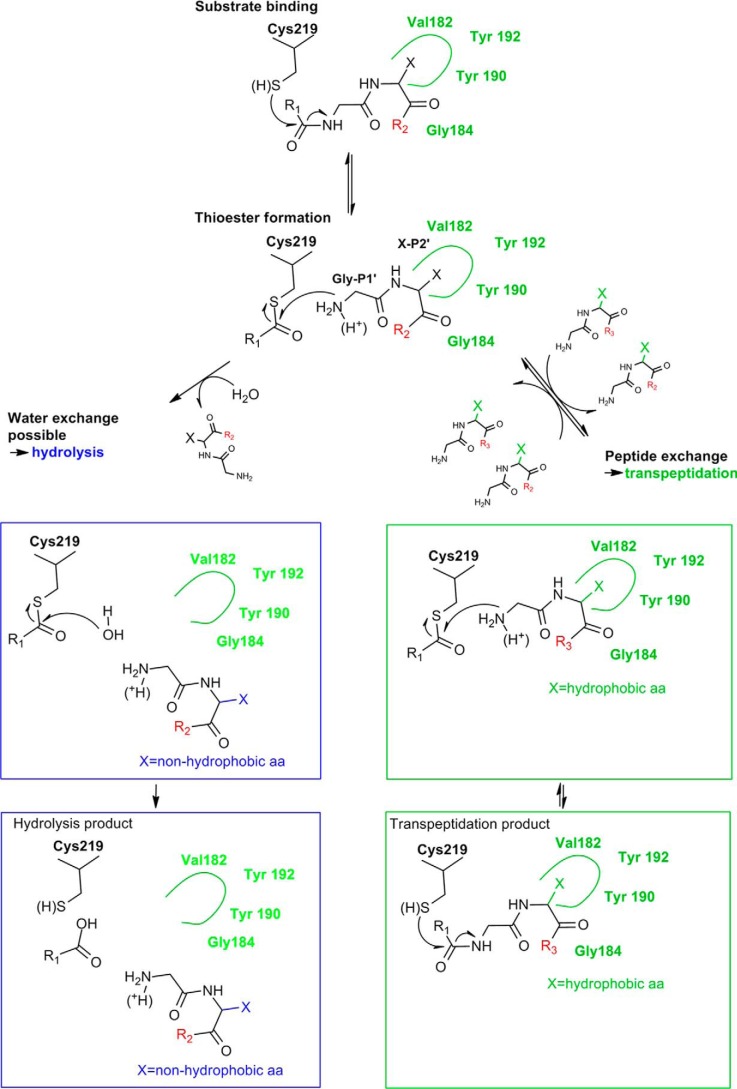
Figure 8.
Mechanistic scheme of transpeptidation and hydrolysis. Green rectangles show transpeptidation reactions induced by a hydrophobic residue in P2′. The hydrolysis reaction pathway is indicated by cyan rectangles, induced by a nonhydrophobic residue in P2′. Starting point: green dashed rectangle. A substrate (R1 + R2) binds to the active site of AtLEGγ. The attack of Cys219 creates a thioester and free N terminus at the primed site. Left pathway, if the initially bound substrate (green dashed line) carries a nonhydrophobic residue in P2′, the primed product can dissociate after the formation of the thioester and water will exchange. Consequently, this results in hydrolysis of the thioester and the release of the hydrolysis product. Right pathway, if P2′ is hydrophobic, the primed site peptide stays bound and prevents the exchange of catalytic water, resulting in an equilibrium between thioester and peptide bond. In the presence of a suitable transpeptidation substrate (R3), an exchange between the initially bound primed product and the transpeptidation peptide can happen. This results in a new equilibrium between thioester and peptide bond, forming the transpeptidation product. The varying protonation state of the released primed N terminus is indicated. Only a deprotonated N terminus is able to attack the thioester, not a protonated one. This relationship explains the pH dependence of transpeptidation, which is more efficient at neutral pH than acidic pH.