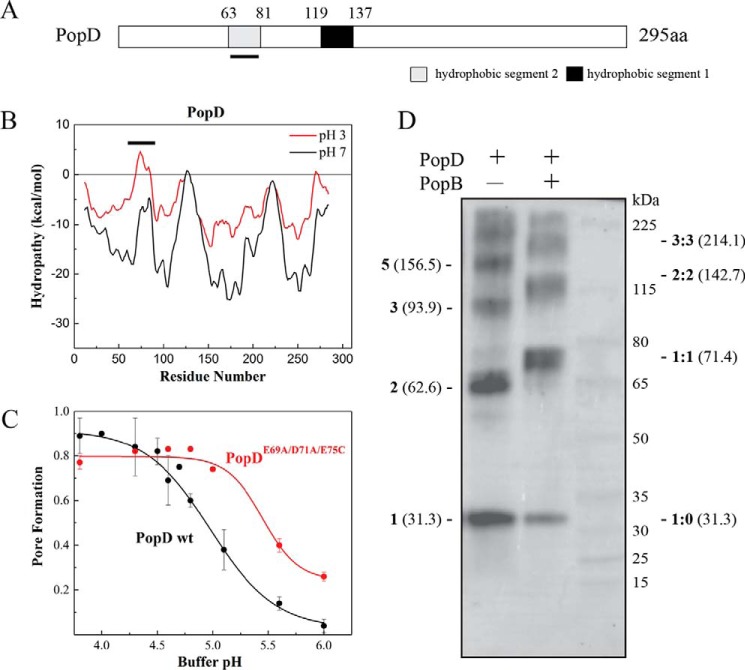
Figure 1.
Identification of a second potential transmembrane segment after protonation of acidic residues in PopD. A, schematic of the primary structure of P. aeruginosa T3S translocator PopD. Predicted hydrophobic segments 1 (H1) and 2 (H2) are shown. The segment H2, whose hydrophobicity increased upon protonation of acidic residues, is underlined. B, hydropathy plot of PopD before (black) and after neutralization of acidic residues (red). The graph was generated using membrane protein explorer (MPEx) with a 19-amino acid sliding window. ΔG was defined as the free energy to transfer amino acids from lipid bilayer to water. The position of segment H2 is indicated with a line. C, pore formation activity of WT PopD and PopDE69A/D71A/E75C at the indicated pH. Pore formation was determined as the fraction of encapsulated Tb(DPA)33− quenched by EDTA as described previously (37). PopD was incubated with liposomes at 20–23 °C for 20 min. The protein:lipid ratio was 1:1000. The mean from two independent experiments are reported with error bars corresponding to the range. D, glutaraldehyde cross-linking of PopD homo-oligomers and PopB and PopD hetero-oligomers formed on liposomes. PopD alone or premixed with an excess of PopB were incubated with liposomes in buffer B for 20 min with a protein:lipid ratio of ∼1:5000. Proteoliposomes were pelleted and subjected to immunoblotting using an anti-PopD antibody. Expected molecular masses for PopD n-mers (left) and PopD:PopB n-mers (right) were estimated using the molecular mass for PopD (31.3 kDa) and PopB (40.1 kDa) monomers.