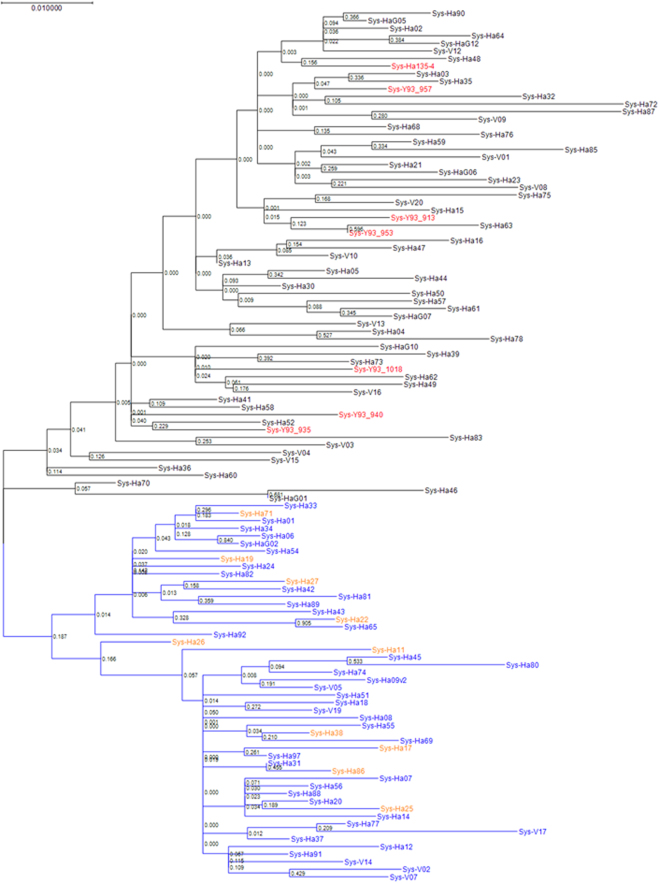
Figure 5.
Phylogenetic Analysis in the NS5B region of Genotype 1b HCV. The genetic distance was estimated using Kimura 2 parameters, and a phylogenetic tree was constructed based on the nucleotide sequences in the NS5B region using the maximum likelihood method adapted with best fit model (Kimura 2-parameter plus Gamma distributed with Invariant sites). The bootstrap values are indicated at each tree root. Sequences with A218 and C316 wild-type (n = 40), A218S mutant and C316 wild-type (n = 10), and A218S and C316N mutant HCV strains (n = 59) observed in 109 sofosbuvir (SOF)-naïve patients are shown in blue, orange and black, respectively. Also, HCV strains (n = 7) detected in patients experiencing virologic failure after SOF-based therapy are shown in red.