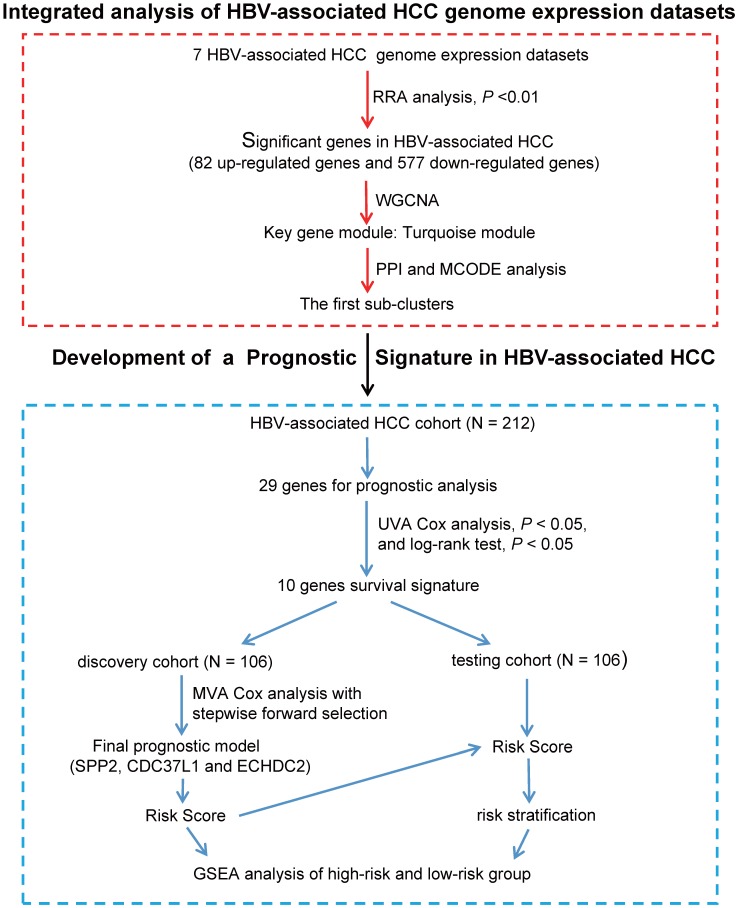
Figure 1.
Flowchart describing the schematic overview of the study design. After integrated analysis and different bioinformatics analysis of HBV-associated HCC genome expression datasets, we identified hub genes in Cluster 1. Hub genes were then analyzed individually for prognostic significance by univariate Cox proportional hazards and Kaplan-Meier survival analysis. 10 hub genes were significantly associated with the survival of patients with HBV-associated HCC (UVA Cox analysis, P < 0.05, and log-rank test P < 0.05). The HBV-associated HCC cohort (GSE14520, N = 212) were randomly divided in to discovery cohort (N = 106) and internal testing cohort (N = 106). Next, we used multivariable Cox proportional hazards stepwise regression analysis with forward selection to build a prognostic model that included 3 genes: SPP2, ECHDC2, and CDC37L1. This model was used to calculate risk scores for discovery cohort (risk score = expSPP2* - 0.1941 + expCDC37L1* - 0.5466 + expECHDC2* - 0.4714), and the cut-off point was chosen. This risk score calculation and cut-off point were further validated in internal testing cohort. Lastly, GSEA analysis of the high-risk and low-risk group was used to further inquiry the 3 genes prognostic signature. UVA Cox: univariate Cox; MVA Cox: multivariate Cox.