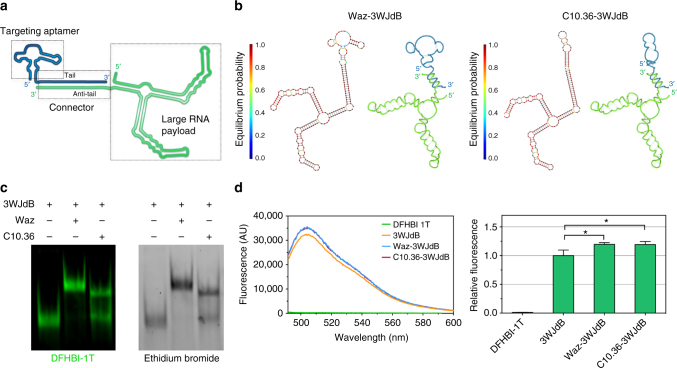
Fig. 1.
Design and assembly of cell-targeting and light-up aptamers. a Schematic representation of the aptamer nanostructure highlights two aptamer modules (targeting and payload) annealed via a connector domain. b Secondary structure predictions of Waz–3WJdB and C10.36–3WJdB. For each aptamer–aptamer hybrid, the predicted hybridization of the two modules based on NUPACK calculations is reported on the left, and a helical depiction is shown on the right. c Non-denaturing gel electrophoresis and dual staining with DFHBI-1T and ethidium bromide show effective formation of Waz–3WJdB and C10.36–3WJdB, and retention of the light-up properties of 3WJdB upon hybridization. Uncropped gel is shown in Supplementary Fig. 1. d Fluorescence spectroscopy in solution of free 3WJdB, Waz–3WJdB, and C10.36–3WJdB (λex = 472 nm; λem = 492–600 nm). Fluorescence emission of 3WJdB (0.5 μM) was measured upon refolding in a buffer supplemented with DFHBI-1T (20 μM) either in the absence or presence of 3-fold molar excess of cell-targeting aptamers relative to 3WJdB. All curves are represented as averages of five independent experiments. Note that blue and magenta curves are overlapping. To calculate the relative fluorescence, we first integrated the fluorescence area from 495 to 600 nm of each sample, and then normalized for the fluorescence of free 3WJdB over this range. Significance was analyzed by one-way ANOVA with post hoc Tukey’s test (*p < 0.05)