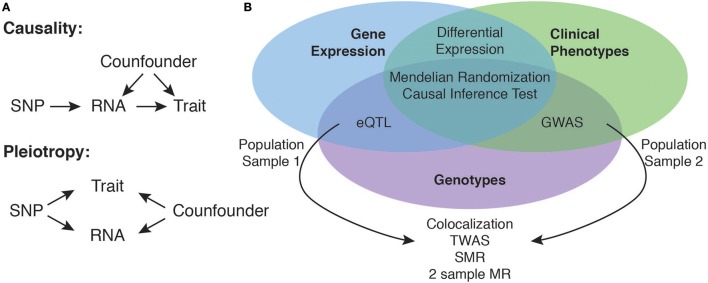
Figure 1.
Using eQTL data to identify causal candidate genes at GWAS loci. Integration of eQTL and GWAS data allows for the identification of candidate causal genes, where the effect of the genetic variant (SNP) on the complex trait is mediated by expression levels of an RNA encoded at the locus (A). Overlapping associations of gene expression and clinical trait at the same locus are however not sufficient to infer causality, as they might also be explained as independent pleiotropic effects (A). Depending on the availability of overlapping individual level data sets of genotypes, gene expression and clinical traits there exist several statistical methods to perform causal inference from the data (B).