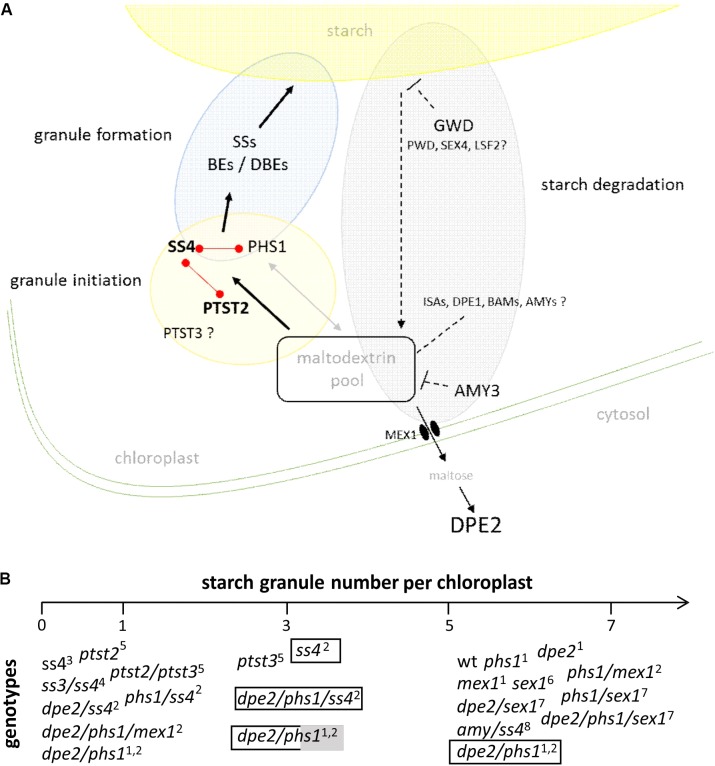
FIGURE 2.
(A) Schematic presentation of starch granule synthesis. For starch granule initiation maltodextrins from the chloroplastidial maltodextrin pool are used by protein targeting to starch 2 (PTST2) and starch synthase 4 (SS4) to produce a granule initiation complex. A lack of the related enzymes massively influences the starch granule number. Enzymes related to initiation reveal protein-protein interaction as depicted by red links. The granule initiation complex is processed by SSs, branching (BEs), and debranching enzymes (DBEs) and a starch granule is formed. The chloroplastidial maltodextrin pool is affected by enzymes acting on glucans, efflux of carbohydrates to cytosol, and influx of carbohydrates via starch degradation. Thus, evidences were published for the involvement of glucan, water dikinase (GWD), disproportionating enzyme 2 (DPE2), α-amylase3 (AMY3), and plastidial phosphorylase (PHS1) during starch granule synthesis. For other enzymes acting on glucans (labeled with ?) the impact on the starch granule synthesis is lacking so far. ISAs- isoamylases; DPE1 and DPE2– disproportionating enzyme 1 and 2; BAMs – β-amylases; AMYs – α-amylases; PWD- phosphoglucan, water dikinase, SEX4 – starch excess 4, LSF2 – like starch excess4, MEX1 – maltose exporter 1 (see also Fettke et al., 2012a). Dashed arrow represents multiple enzymatic reactions. (B) Starch granule number per chloroplast in different genotypes. Plants were grown under 12 h ligth/12 h dark unless otherwise specified: transparent bar – continuous light, transparent/grey bar – 14 h light, 10 h dark.1Malinova et al., 2014; 2Malinova et al., 2017; 3Roldán et al., 2007; 4D’Hulst and Mérida, 2012; 5Seung et al., 2017; 6Mahlow et al., 2014; 7Malinova and Fettke, 2017; 8Seung et al., 2016.