Abstract
Syringa oblata Lindl. (S. oblata) has been used in herbal medicines for treating bacterial diseases. It is also thought to inhibit Streptococcus suis (S. suis) biofilm formation. However, due to the inherent nature of the complexity in its chemical properties, it is difficult to understand the possible bioactive ingredients of S. oblata. The spectrum-effect relationships method was applied to screen the main active ingredients in S. oblata obtained from Heilongjiang Province based on gray relational analysis. The results revealed that Sub-MICs obtained from 10 batches of S. oblata could inhibit biofilm formation by S. suis. Gray relational analysis revealed variations in the contents of 15 main peaks and rutin was discovered to be the main active ingredient. Then, the function of rutin was further verified by inhibiting S. suis biofilm formation using crystal violet staining. Computational studies revealed that rutin may target the chloramphenicol acetyltransferase protein in the biofilm formation of S. suis. In conclusion, this study revealed that the spectrum-effect relationships and computational studies are useful tools to associate the active ingredient with the potential anti-biofilm effects of S. oblata. Here, our findings would provide foundation for the further understanding of the mechanism of S. oblata intervention in biofilm formation.
Keywords: Syringa oblata Lindl., spectrum-effect relationships, active ingredients, biofilm formation, computational studies
Introduction
To date, herbal medicines are gaining more and more attention in recent years because of the availability of multiple chemical ingredients (Yi et al., 2014). Among others, chemical fingerprint analysis has the potential of characterizing the key components and unknown components in a complex system (Lee et al., 2009). Nevertheless, it is very difficult to determine the main ingredients that provide the therapeutic effects in herbal medicines (Kong et al., 2011). It is well-known that spectrum-effect relationships analysis (Kong et al., 2009b) and chemical fingerprints have been used to determine the main ingredients in herbal medicines. In addition, spectrum-effect relationships which applied to the screening and analysis of active ingredients were characterized by similarity analysis, clustering analysis, principal component analysis (PCA), gray relational analysis, partial least squares, and so on (Shi et al., 2016). Among them, gray relational analysis has been certified to be a useful tool to deal with poor, incomplete, and uncertain information (Kuo et al., 2008). The gray system theory can be used to solve the complicated interrelationships among the multiple performance characteristics effectively (Lin and Lin, 2002). Thus, similarity analysis, clustering analysis, PCA and gray relational analysis may be able to elucidate the association between therapeutic effect and main component in herbal medicines.
To find the main ingredients, we chose Syringae Folium leaves as samples. Syringae Folium leaves are mainly from the dry leaves of Oleaceae plant: Syringa oblata Lindl. (S. oblata), Syringa diatata Nakai, and Syringa vulgaris L. Among them, S. oblata is widely distributed in Heilongjiang Province and has been proved to have a huge potential in the development of Chinese medicine through its plant resources, chemical constituents, pharmacological action, and clinical application (Bai et al., 2017). It is well-distributed in the temperate and frigid temperate regions of the middle and high latitudes of Eurasia and North America (Liu et al., 2010). There are also more than 20 species in China, beginning from the northeast to the southwest provinces (Su et al., 2015). Among the Syringae spp. found in the temperate regions of Heilongjiang Province, S. oblata is widely cultivated. It has strong vitality and complex chemical composition. It contains syringopicroside (Liu et al., 2010) and oleuropein (Bi et al., 2011). There are also other constituents such as flavonoids, e.g., rutin (Wang S. et al., 2017), as well as volatile oil compounds (Venturini et al., 2014).
In addition, the reports have shown that S. oblata has significant pharmacological properties such as anti-bacterial (Bai et al., 2017) and anti-inflammatory (Liu and Wang, 2011). It has also been reported that sub-MICs of aqueous extracts of S. oblata Lindl. decreases biofilm formation by Streptococcus suis (Bai et al., 2017). Using gray relational analysis, we could identify the main ingredients. Furthermore, Bai et al. (2017) have predicted that the up-regulated chloramphenicol acetyltransferase (CAT) protein of S. suis was possibly associated with biofilm formation (McLennan et al., 2008; Sahni et al., 2009). However, the detailed molecular interaction between the main ingredient and the 3D CAT protein is still unknown. Currently, computer docking analysis is an internationally recognized method employed in interpreting the relationship between bioactive ingredients and their perceived therapeutic effects (Mordalski et al., 2015; Haddad et al., 2016). Molecular docking is performed to predict the interaction between compounds and protein (Tulahong, 2015). If the 3D structure of CAT protein is constructed, we may identify the interaction between compounds and CAT protein by docking studies. It would lay the foundation for understanding the molecular mechanism involved in the inhibition of biofilm formation in S. suis by the main ingredient.
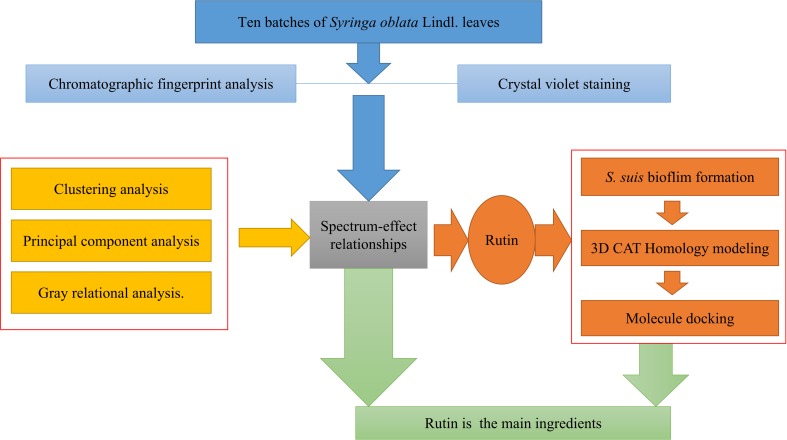
In this study, according to the established fingerprint, 10 batches of S. oblata collected from different regions (Heilongjiang Province, China) were analyzed and the main ingredients of S. oblata were screened by gray relational analysis. The main ingredient was verified by inhibiting the biofilm formation of S. suis in vitro and the docking-based virtual screening. Above all, the study offers a new approach for the determination of the active ingredients in S. oblata and the most effective substance against S. suis biofilm formation. The experimental flow chart of this study is shown in Figure 1.
FIGURE 1.
The experimental flow chart.
Materials and Methods
Plant Materials
Ten batches of samples were collected from different regions in Heilongjiang Province of China in September, 2015 (Table 1 and Supplementary Material 1), and their species were identified after drying the leaves at room temperature by Professor Mingxia Bai in Heilongjiang Academy of Agricultural Sciences Horticulture Branch. Then the samples were blended and sifted through the 24-mesh sieve.
Table 1.
Representative samples of S. oblata in September, 2015 in Heilongjiang investigated in this study.
| No. | Sample origin | Highest temperature (°C) | Lowest temperature (°C) | Average temperature (°C) |
|---|---|---|---|---|
| S1 | Daqing | 28 | 3 | 17 |
| S2 | Jiagedaqi | 23 | -3 | 12 |
| S3 | Harbin | 26 | 4 | 16 |
| S4 | Hegang | 28 | 0 | 15 |
| S5 | Jixi | 27 | 2 | 15 |
| S6 | Jiamusi | 27 | 1 | 16 |
| S7 | Qiqihaer | 28 | 3 | 17 |
| S8 | Shuangyashan | 26 | 1 | 15 |
| S9 | Mudanjiang | 28 | -1 | 16 |
| S10 | Zhaodong | 28 | 2 | 17 |
Chemical Analysis
Chromatographic fingerprint analysis was done using Waters Alliance HPLC system (Shimadzu, Corporation, Kyoto, Japan) that is equipped with a binary pump and a UV/V is detector. The chromatographic separation was performed on a Diamosil C18 column (4.6 mm × 250 mm, 5 μm) using aqueous solution of 0.1% formic acid (solvent A) and acetonitrile (solvent B) as mobile phase at a flow rate of 1 mL/min. The gradient program was 0 min, 5% solvent B; 30 min, 53% solvent B; and 35 min, 5% solvent B. The chromatogram was monitored at a wavelength of 245 nm during the experiment. The method was optimized by precision, repeatability, and stability of the target on the relative retention time and relative peak area in Supplementary Material 2.
All samples were completely dried at room temperature. Ten batches of samples of S. oblata leaves (leaves, 200 g) were boiled in deionized water (10 times of the amount of leaves) for 45 min, decanted, filtered and evaporated at 60°C. Aqueous extract of these dried S. oblata leaves was weighed and reconstituted to a concentration of 3 mg/mL. The samples were filtered through a 0.45 μm membrane filter prior to HPLC analysis (Bai et al., 2017).
Thereafter, all samples were analyzed by Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine (Version 2004A) (Kong et al., 2009b) which is recommended by State Food and Drug Administration of China (SFDA). Clustering analysis and PCA are also very popular visualization tools in analyzing the samples and these were done by the software Metabo Analyst 3.0. Gray relational analysis which is a multi-factor statistical analysis method (Zhang and Zhang, 2007) was used to determine the correlation between the 15 common peaks of the fingerprint of each S. oblata leaf sample and the data obtained from the inhibition of S. suis biofilm. Using spectrum-effect analysis, fingerprinting, and biofilm formation on the 10 batches of S. oblata samples were analyzed. The experiment used “Gray Correlation Software (Version 3)” to calculate gray relational degree.
Bacterial Strains and Culture Conditions
In this study, S. suis ATCC700794 strain was cultured in Todd Hewitt broth (THB) (THB: Summus, Ltd., Harbin, Heilongjiang, China) and Todd Hewitt broth agar (THA) supplemented with 5% (v/v) fetal bovine serum (Sijiqing, Ltd., Hangzhou, Zhejiang, China) at 37°C. The original THB was supplemented with 1.8% Bacto-agar (Difco Laboratories) in order to obtain THA. All the media were autoclaved for 30 min at 121°C. The strain was transferred from the stock cultures to THA prior to inoculation and incubated aerobically at 37°C for 16 h. Then, the strain was repeatedly subcultured under the same conditions. The final cultures were used for the minimum inhibitory concentration (MICs) assays and the biofilm formation assays.
Determination of Minimum Inhibitory Concentration (MIC)
The MICs of the 10 batches of dried S. oblata samples and rutin standard solution of methanol (Guoyao, Ltd., China) were determined by Microdilution method (Yang et al., 2015). Overnight cultures of S. suis were diluted in sterile saline to a density of McFarland 0.5 standard which corresponds to 1 × 108 colony-forming units (CFU)/mL. Thereafter, the cultures of S. suis were further diluted to 1:100 using sterile THB. Then 100 μL each of the samples and rutin were added to each well of a 96-well plate (Corning Costar® 3599, Corning, NY, United States) in 100 μL culture medium, respectively. Control bacteria were cultivated in the absence of the 10 batches of dried S. oblata samples and rutin. After the incubation for 24 h at 37°C, the MICs were determined as the lowest concentration of the 10 batches of S. oblata samples and rutin.
S. suis Biofilm Formation Assays
The S. suis biofilm formation was evaluated using crystal violet staining (Corning Costar® 3599, Corning, NY, United States) (Yang et al., 2015). S. suis was grown in a Costar R 3599 96-well plate in the presence of the 10 batches of 1/2 MIC of S. oblata samples and 1/2 MIC, 1/4 MIC, and 1/8 MIC of rutin. Negative and positive control wells contained culture medium and bacterial suspension, respectively. Biofilms were treated as described previously (Yang et al., 2015). The plates were incubated for 24 h at 37°C. Then, the remaining bacteria were aspirated and washed three times with 0.25 mL of sterile physiological saline. The remaining attached bacteria were fixed with 0.2 mL of 99% methanol (Guoyao, Ltd., China) per well in 15 min and 0.2 mL of 2% crystal violet (Guoyao, Ltd., China). After the plates were air dried, the dye was resolubilized with 0.2 mL of 33% (v/v) glacial acetic acid (Guoyao, Ltd., China) per well. Using a microtiter plate reader (DG5033A, Huadong, Ltd., Nanjing, Jiangsu, China), the OD of each well was measured at 570 nm.
Computational Studies
In order to determine the main ingredients of S. oblata samples targeting the formation of S. suis biofilm, we constructed the 3D structure of CAT. Although there was no available crystal structure of the CAT protein from S. suis, the amino acid sequence of CAT was retrieved from UniProt (Accession No. C6GT52). It is important to construct the 3D structure based on the known structures by homology modeling method as described previously (Chen et al., 2017). Then, a series of homology methods including sequence similarity searches, target sequences were constructed. The total energy of lowest probability density functions (PDFs) and discrete optimized potential energy (DOPE) score were performed in order to construct an appropriate 3D model of CAT protein of S. suis using Discovery Studio (version 3.0). Thereafter, the best model was assessed and verified. Finally, molecular docking calculations were performed with DS-LibDock protocol under default conditions (Chen et al., 2017).
Statistical Analysis
All the experiments were performed in triplicates. Data analysis and calculation of standard deviation were done using SPSS 11.0.0 (IBM, United States).
Results
HPLC Fingerprints of S. oblata Samples
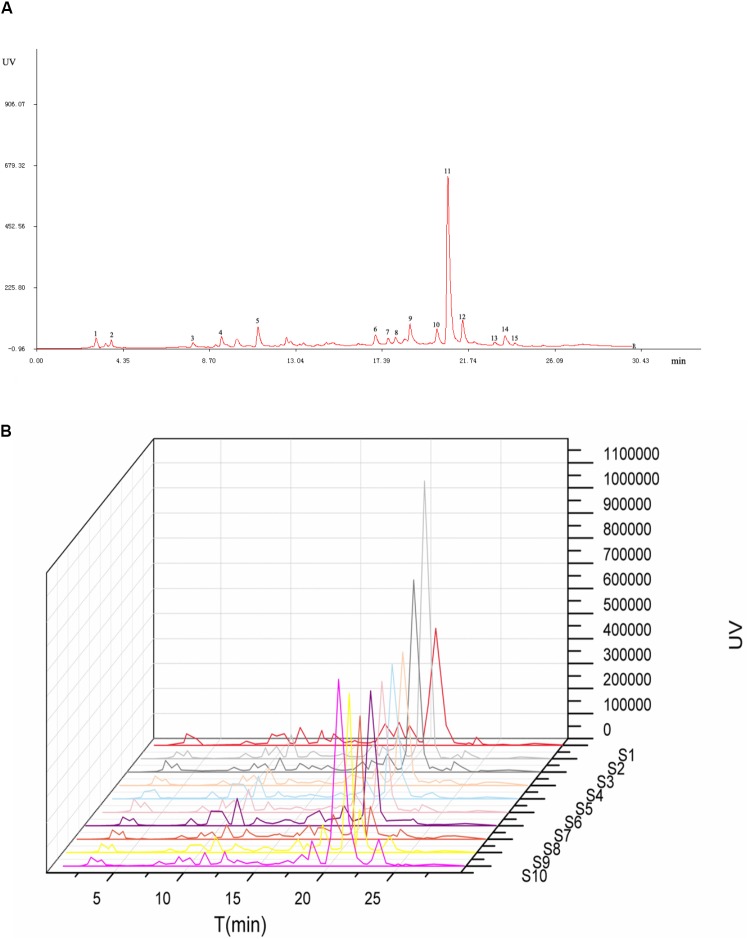
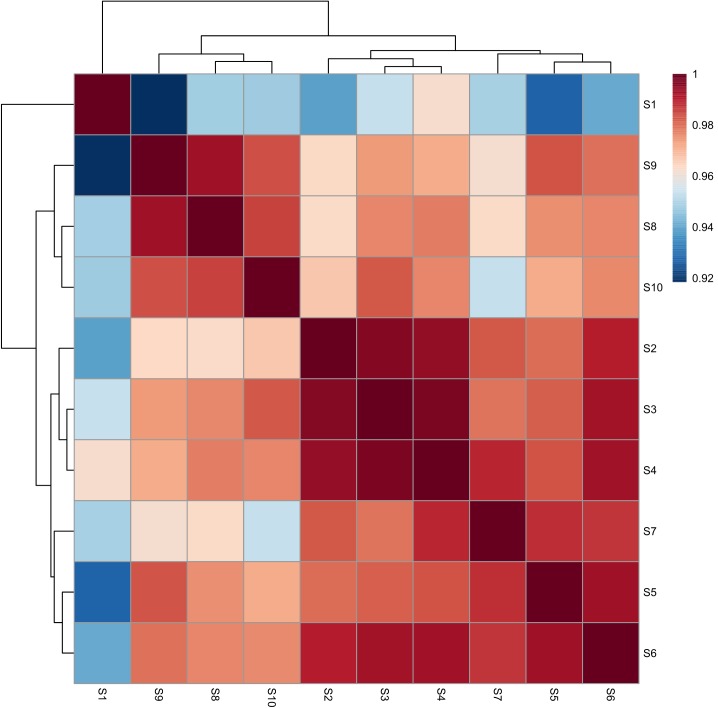
The HPLC fingerprints of S. oblata samples are shown in Figure 2. The results revealed that there were differences in the fingerprints among the 10 batches of S. oblata samples (Figure 2B). Thereafter, the professional similarities were calculated using the Similarity Evaluation System for Chromatographic Fingerprint of herbal medicines (Version 2004A) (Kong et al., 2009b). Similarities between the reference fingerprint (Figure 2A) and each chromatographic profile of the 10 batches of S. oblata samples (Figure 2B) were evaluated by calculating the correlation coefficient in Figure 3. These results indicated that S1 and S2 which were located in oil field with poor soil conditions and a low temperature region respectively had the lowest degree of similarity (Liao et al., 2015). This may be responsible for the differences in the fingerprints of S. oblata. Despite this, these results revealed that the 10 batches of samples of S. oblata from different locations shared nearly the same correlation coefficients (over 0.92) of similarities and similar chemical constituents.
FIGURE 2.
Chromatographic fingerprints for reference (A) and all the Syringa oblata sample (B).
FIGURE 3.
The correlation coefficient for all the S. oblata sample.
Clustering Analysis
Clustering analysis is one of the detection and quantification classification evaluation methods that is commonly used in analyzing samples of herbal medicines (Kong et al., 2009a). Clustering analysis method is well-known and has gained wide range application in fingerprint analysis. This is because its application is simple and its method of data interpretation is non-parametric (Zou et al., 2005). The 10 samples were analyzed using clustering analysis (Dong et al., 2017) of the peak areas of the 15 common analytical markers in the HPLC fingerprints. Results of cluster analysis showed that the fingerprints separated into seven clusters, as shown in Figure 4A. Cluster 1 contained only S10; cluster 2 included S8 and S9; cluster 3 included only S1; cluster 4 included S5 and S7; cluster 5 contained only S2; cluster 6 contained only S6 and cluster 7 contained S3 and S4. These results could be explained as follows: the differences in the chemical ingredients could be related to the different geographic locations. Interestingly, the results of clustering analysis were consistent with those of similarity analysis.
FIGURE 4.
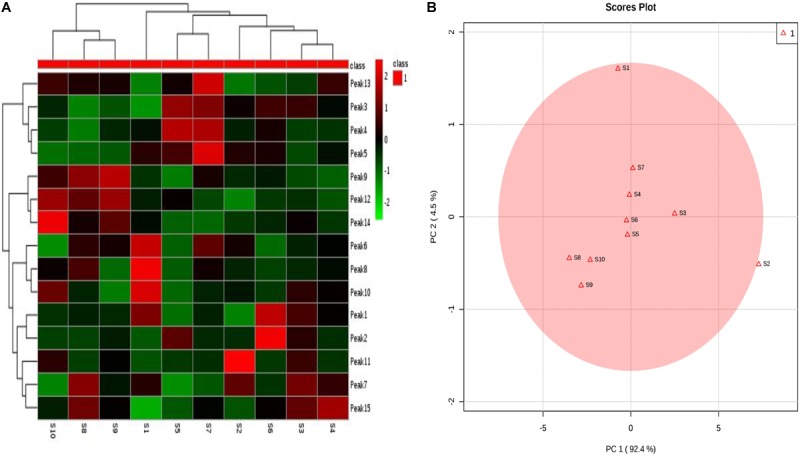
Syringa oblata samples from different sources: (A) heatmaps of 15 constituent content; (B) principal component analysis (PCA) results.
Principal Component Analysis
In addition, the results of PCA are presented in Figure 4B. The first two principal components (PC1 and PC2) revealed the clear differences in the cultivating locations, which accounted for the 92.4% and 4.5% of the total variability. From Figure 4B, it could be seen that the samples were clearly divided into five groups: group 1 (S4, S5, S6, S7); group 2 (S8, S9, S10); group 3 (S1); group 4 (S2); and group 5 (S3). PCA could further be used to evaluate the information of the fingerprints and provide reference to assess the quality of different batches of S. oblata. The differences in the results revealed that the uniqueness in the compositions of the samples of S. oblata may be due to the different environmental conditions in which the plants were exposed. In particular, it was clear from the map (Supplementary Material 1) that groups 3, 4, and 5 were far apart from groups 1 and 2. Interestingly, S2 came from a low temperature region, S3 was obtained from the capital city of Heilongjiang (with a high-rise building region in the southwest) and S1 was obtained from the south western region (oil field) with poor soil conditions with little difference from other regions, proving the feasibility of PCA. Furthermore, PCA had helped in effectively guiding the study and understanding of the active ingredients in S. oblata. In spite of this, no difference was observed in the results of clustering analysis and similarity analysis among the 10 batches of S. oblata.
Spectrum-Effect Relationship
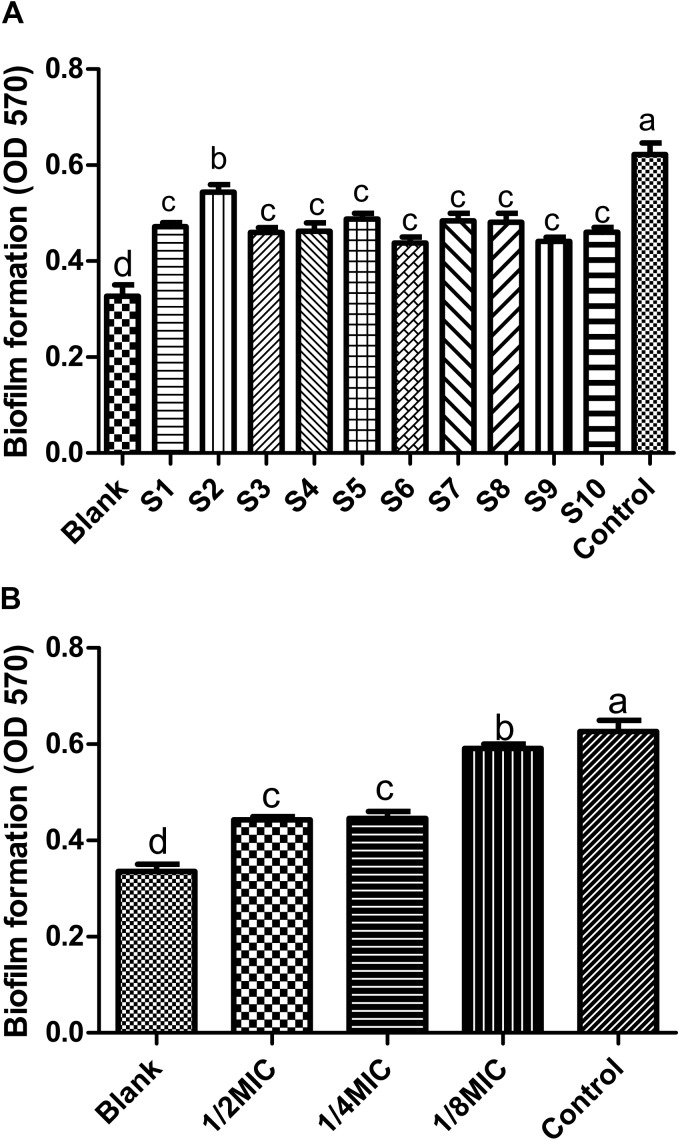
In order to find out the main active ingredients involved in the inhibition of biofilm formation, 10 batches of S. oblata samples were evaluated in S. suis biofilm. The MICs were determined as 12.5 mg/mL, except for S2 and S9 which were determined as 6.25 mg/mL. The results revealed that the 1/2 MIC of 10 batches of the S. oblate samples had the ability to significantly inhibit the formation of S. suis biofilm (p < 0.05) (Figure 5A) compared with positive control. Among them, S2, which was obtained from a low temperature region, was different from other regions in inhibiting the formation of S. suis biofilm. In addition, gray relational analysis was used to determine the correlation between the 15 common peaks of fingerprint and S. suis biofilm data of each S. oblata sample. The results revealed that the characteristic peak 6 was the greatest gray relational rankings in Table 2. It was closely related to S. suis biofilm formation and might be the main pharmaceutical component of S. oblata which inhibited the S. suis biofilm. Previous studies have shown that the main component in characteristic peak 6 was rutin at the retention time of 17.66 min by HPLC-ESI-MS analysis (Bai et al., 2017). Thus, the rutin was evaluated to monitor its effect against biofilm formation by S. suis. The results revealed that the MIC of rutin against S. suis was 0.3125 mg/mL. The inhibitory effect of rutin was significant for 1/2 MIC and 1/4 MIC (p < 0.05), with no effect by 1/8 MIC compared with positive control (Figure 5B). The contents of rutin in 10 batches of S. oblata in MICs are shown in Supplementary Material 2.
FIGURE 5.
Inhibitory effect on biofilm formation by Streptococcus suis (n = 5, x ± SD): (A) 10 batches of S. oblata samples; (B) the rutin standard. Different letters indicate a significant difference at p < 0.05.
Table 2.
Gray relational grade of the peak with biofilm formation.
| No. | Gray relation | Gray relational rankings |
|---|---|---|
| 1 | 0.777193 | 12 |
| 2 | 0.709735 | 13 |
| 3 | 0.881281 | 4 |
| 4 | 0.901884 | 2 |
| 5 | 0.900460 | 3 |
| 6 | 0.905249 | 1 |
| 7 | 0.864141 | 6 |
| 8 | 0.876266 | 5 |
| 9 | 0.859622 | 7 |
| 10 | 0.799613 | 11 |
| 11 | 0.841602 | 9 |
| 12 | 0.682051 | 14 |
| 13 | 0.845761 | 8 |
| 14 | 0.521954 | 15 |
| 15 | 0.896398 | 10 |
Computational Studies
Homology modeling of CAT protein was conducted. The 3D structure of CAT from Escherichia coli (PDB code: 3CLAA), Escherichia coli (PDB code: 1Q23L) and Bacteroides thetaiotaomicron (PDB code: 2I9DC) were screened as the template structure. The best homology model was established based on the least PDF total energy (11550.45) and DOPE score (-23049.50). The final model which had the maximum number of residues in the favored regions (93.26%) was selected. Indeed, the Z-score calculated for the template was -1.56. The results demonstrated that the established homology model of the CAT should be a reasonable structure. The results are presented in Supplementary Material 3. Thereafter, the active-site was copied from Escherichia coli (PDB code: 3CLAA). Finally, the results revealed that rutin could interact with CAT protein having a LibDock score (100.31) by docking analysis. From the results, we observed that rutin targets the CAT protein with three hydrogen bonds and two p-π interactions (Figures 6, 7). Above all, we observed that rutin could combine with CAT protein to inhibit biofilm formation by S. suis.
FIGURE 6.
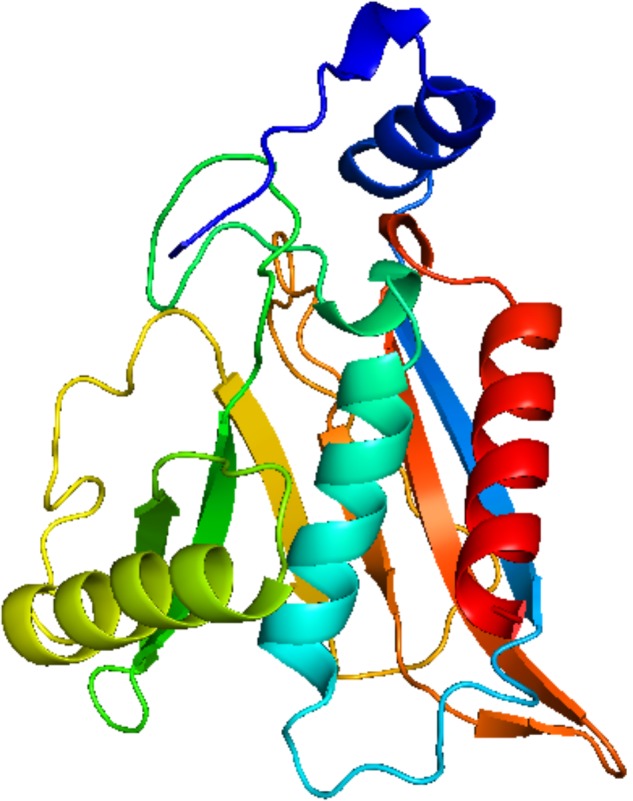
Modeled structure of CAT protein from Streptococcus suis.
FIGURE 7.
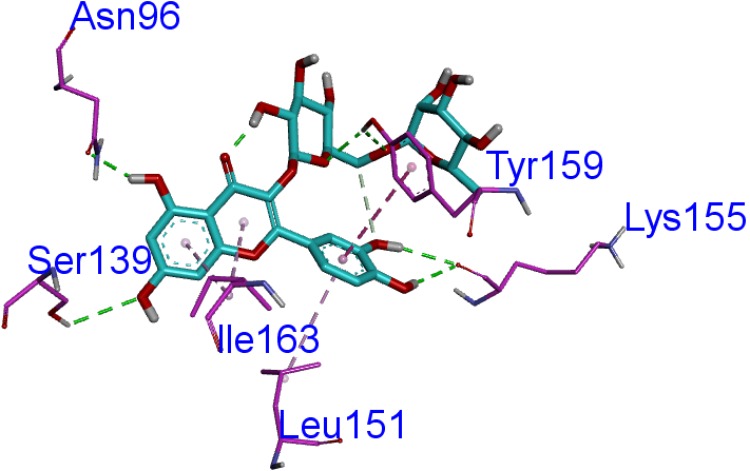
The ligand of rutin compared by the LibDock docking method. The atoms of the compound, rutin, are shown in ball-and-stick format and colored by atom. Polar interactions are shown as dashed colored lines. Among them, the green dotted line represents the hydrogen bond interaction. The pink dotted line represents p-π interactions interaction.
Discussion
Herbal medicines have been widely used for treating bacterial diseases (Wong et al., 2010). It has a long therapeutic history over thousands of years (Chang et al., 2008; Kong et al., 2009a). S. suis is one of the leading sources of high economic losses in the pig industry worldwide, which causes a wide variety of disease, including meningitis, arthritis, septicemia and bacterial diseases (Wang et al., 2011, 2016). In addition, it is important to control the formation of S. suis biofilm in the fight against bacterial diseases in pigs (Wang et al., 2012). Recent studies have shown that Leptospermum scoparium (manuka) (Song et al., 2013), Glycyrrhiza (Guldas et al., 2016), and Punica granatum L. plants (Bakkiyaraj et al., 2013) can also inhibit biofilm formation. Furthermore, it has also been reported that aqueous extract of sub-MICs of S. oblata Lindl. aqueous extract decreased biofilm formation in S. suis (Bai et al., 2017). In this study, a crystal violet staining (Yang et al., 2015) was used to evaluate the anti-biofilm effects of S. suis. The results indicated that 1/2 MIC derived from 10 batches of dried S. oblata samples significantly decreased the biofilm formation capability of S. suis. This is in consonance with previous study (Bai et al., 2017). However, this plant material is made up of complex chemical composition. Using chemical fingerprints, it was difficult to determine active ingredient in S. oblata that was responsible for inhibiting biofilm formation among the chemical constituents (Wang Y. et al., 2017).
Thus, we used spectrum-effect relationships analysis (Kong et al., 2009b) to characterize the bioactive substance in S. oblata samples that inhibited S. suis biofilm formation. First, the results revealed that the combination of similarity analysis, clustering analysis, and PCA was able to effectively classify the samples objectively in accordance with different factors, such as geographical locations, elevation and the climatic conditions they were exposed to. Although the chemical fingerprints could show the differences between most of the constituents, it could not directly show the anti-biofilm effect of S. oblata. Thus, the gray relational analysis combined with the study of the effect of the chemical components on biofilm formation was used to obtain the bioactive ingredients. At the same time, gray relational analysis could explore the correlation of fingerprints and the anti-inflammatory effects directly (Lu et al., 2017) and be used to evaluate the multiple performance characteristics of the bioactive substance and then be optimized into a single gray relational grade (Lin and Lin, 2002). Thus, it was analyzed with S. suis biofilm data of each S. oblata sample and the peak area of 15 constituents. In our study, the characteristic peak 6 played an important role in the inhibition of S. suis biofilm formation. This proved that rutin, which corresponded to the characteristic peak 6, was the potential substance that inhibited the formation of S. suis biofilm, and this finding is in agreement with earlier findings previously reported by Bai et al. (2017). There is a direct relationship between the content of rutin and its ability to inhibit S. suis biofilm. Kong et al. (2017) also reported that the most abundant compounds might not be the major contributors to antibacterial activity. This is also in tandem with our study. However, it was unclear how rutin inhibited S. suis biofilm formation.
Previous study has reported that some differentially expressed proteins may be involved with the formation and inhibition of biofilm by S. suis for relative and absolute quantitation (iTRAQ) labeling (Bai et al., 2017). It was discovered that the up-regulated CAT protein may inhibit the biofilm information mechanism of bacteria (McLennan et al., 2008; Sahni et al., 2009). The decreased levels of acetyl coenzyme A (acetyl-CoA) (Potrykus and Wegrzyn, 2001) was shown in CAT-expressing cells and acetyl-CoA played a role in biofilm formation (Pruess et al., 2010). Thus, in order to discover the active ingredients of S. oblata responsible for the mechanism involved in the inhibition of biofilm formation by S. suis, docking calculations were performed with DS-LibDock protocol using DS 3.0, which revealed that rutin could interact well with CAT protein which was built by homology modeling. Then the active-site was copied from Escherichia coli (PDB code: 3CLAA). Finally, the results from docking analysis proved that rutin could interact with CAT protein with LibDock score (100.31). The rutin attracted the CAT protein in S. suis through specific residual interaction with three hydrogen bonds of ASN 96, TYR 159, and SER 139 of CAT. In addition, there were also two p-π interactions of ILE 163 and LEU 151 with CAT acid side-chains (Figures 6, 7). It may be possible to enhance CAT thermostability via a rational design approach and optimize the amino acid configuration surrounding (Kim et al., 1997). Our results speculate that rutin can also enhance the stability of the CAT and inhibit S. suis biofilm. Interestingly, this is the first report that rutin targets the CAT protein in S. suis biofilm.
Conclusion
Ten batches of S. oblata from Heilongjiang province were analyzed using chemical fingerprint analysis. Then, spectrum-effect relationships confirmed rutin as the main bioactive ingredient in S. oblate which inhibits biofilm formation by S. suis. Molecular docking revealed that rutin targeted the CAT protein in S. suis biofilm thus leading to its inhibition. The novelty of this work is the identification of rutin as the active ingredient of S. oblata by spectrum-effect relationships and computational studies. Our findings here would provide the foundation for the further understanding of the mechanism of S. oblata intervention in biofilm formation.
Author Contributions
Y-HL designed the whole experiment. Y-YL and X-RC directed the completion of the experiment. L-FG, MC, W-QC, W-YD, X-YC, and B-OG were supportive during the experiment.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (Grant No. 31472231) and the earmarked fund for China Agriculture Research System -35.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2018.00570/full#supplementary-material
References
- Bai J., Yang Y., Wang S., Gao L., Chen J., Ren Y., et al. (2017). Syringa oblata Lindl. Aqueous Extract Is a Potential Biofilm Inhibitor in S. suis. Front. Pharmacol. 8:26. 10.3389/fphar.2017.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkiyaraj D., Nandhini J. R., Malathy B., Pandian S. K. (2013). The anti-biofilm potential of pomegranate (Punica granatum L.) extract against human bacterial and fungal pathogens. Biofouling 29 929–937. 10.1080/08927014.2013.820825 [DOI] [PubMed] [Google Scholar]
- Bi X., Li W., Sasaki T., Li Q., Mitsuhata N., Asada Y., et al. (2011). Secoiridoid glucosides and related compounds from Syringa reticulata and their antioxidant activities. Bioorg. Med. Chem. Lett. 21 6426–6429. 10.1016/j.bmcl.2011.08.089 [DOI] [PubMed] [Google Scholar]
- Chang Y. X., Ding X. P., Qi J., Cao J., Kang L. Y., Zhu D. N., et al. (2008). The antioxidant-activity-integrated fingerprint: an advantageous tool for the evaluation of quality of herbal medicines. J. Chromatogr. A 1208 76–82. 10.1016/j.chroma.2008.08.054 [DOI] [PubMed] [Google Scholar]
- Chen X. R., Wang X. T., Hao M. Q., Zhou Y. H., Cui W. Q., Xing X. X., et al. (2017). Homology modeling and virtual screening to discover potent inhibitors targeting the imidazole glycerophosphate dehydratase protein in Staphylococcus xylosus. Front. Chem. 5:98. 10.3389/fchem.2017.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R. F., Su J., Nian H., Shen H., Zhai X., Xin H. L., et al. (2017). Chemical fingerprint and quantitative analysis of flavonoids for quality control of Sea buckthorn leaves by HPLC and UHPLC-ESI-QTOF-MS. J. Funct. Foods 37 513–522. 10.1016/j.jff.2017.08.019 [DOI] [Google Scholar]
- Guldas H. E., Kececi A. D., Cetin E. S., Ozturk T., Kaya B. U. (2016). Evaluation of antimicrobial efficacy of cetrimide and Glycyrrhiza glabra L. extract against Enterococcus faecalis biofilm grown on dentin discs in comparison with NaOCl. Dent. Mater. J. 35 721–727. 10.4012/dmj.2014-2338 [DOI] [PubMed] [Google Scholar]
- Haddad Y., Heger Z., Adam V. (2016). Guidelines for homology modeling of dopamine, norepinephrine, and serotonin transporters. ACS Chem. Neurosci. 7 1607–1613. 10.1021/acschemneuro.6b00242 [DOI] [PubMed] [Google Scholar]
- Kim S. J., Jeon H. Y., Han B. K. (1997). Chloramphenicol acetyltransferase expression of Escherichia coli is increased at 42°C. Biotechnol. Tech. 11 435–438. 10.1023/A:1018481125858 [DOI] [Google Scholar]
- Kong W. J., Wang J. B., Zang Q. C., Xing X. Y., Zhao Y. L., Liu W., et al. (2011). Fingerprint-efficacy study of artificial Calculus bovis in quality control of Chinese materia medica. Food Chem. 127 1342–1347. 10.1016/j.foodchem.2011.01.095 [DOI] [PubMed] [Google Scholar]
- Kong W. J., Zhang S. S., Zhao Y. L., Wu M. Q., Chen P., Wu X. R., et al. (2017). Combination of chemical fingerprint and bioactivity evaluation to explore the antibacterial components of Salvia miltiorrhizae. Sci. Rep. 7:8112. 10.1038/s41598-017-08377-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W. J., Zhao Y. L., Xiao X. H., Jin C., Li Z. L. (2009a). Quantitative and chemical fingerprint analysis for quality control of Rhizoma Coptidischinensis based on UPLC-PAD combined with chemometrics methods. Phytomedicine 16 950–959. 10.1016/j.phymed.2009.03.016 [DOI] [PubMed] [Google Scholar]
- Kong W. J., Zhao Y. L., Xiao X. H., Wang J. B., Li H. B., Li Z. L., et al. (2009b). Spectrum-effect relationships between ultra performance liquid chromatography fingerprints and anti-bacterial activities of Rhizoma coptidis. Anal. Chim. Acta 634 279–285. 10.1016/j.aca.2009.01.005 [DOI] [PubMed] [Google Scholar]
- Kuo Y., Yang T., Huang G. W. (2008). The use of grey relational analysis in solving multiple attribute decision-making problems. Comput. Ind. Eng. 55 80–93. 10.1016/j.cie.2007.12.002 [DOI] [Google Scholar]
- Lee M. K., Ahn Y. M., Lee K. R., Jung J. H., Jung O.-S., Hong J. (2009). Development of a validated liquid chromatographic method for the quality control of Prunellae Spica: determination of triterpenic acids. Anal. Chim. Acta 633 271–277. 10.1016/j.aca.2008.12.038 [DOI] [PubMed] [Google Scholar]
- Liao J., Wang J., Jiang D., Wang M. C., Huang Y. (2015). Long-term oil contamination causes similar changes in microbial communities of two distinct soils. Appl. Microbiol. Biotechnol. 99 10299–10310. 10.1007/s00253-015-6880-y [DOI] [PubMed] [Google Scholar]
- Lin J. L., Lin C. L. (2002). The use of the orthogonal array with grey relational analysis to optimize the electrical discharge machining process with multiple performance characteristics. Int. J. Mach. Tools Manuf. 42 237–244. 10.1016/S0890-6955(01)00107-9 [DOI] [Google Scholar]
- Liu X., Wang J. (2011). Anti-inflammatory effects of iridoid glycosides fraction of Folium syringae leaves on TNBS-induced colitis in rats. J. Ethnopharmacol. 133 780–787. 10.1016/j.jep.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Liu X., Wang J. M., Zhou C. X., Gan L. S. (2010). Preparative separation and enrichment of syringopicroside from Folium syringae leaves with macroporous resins. J. Biomed. Biotechnol. 2010:572570. 10.1155/2010/572570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S. W., Dong S., Xu D., Duan J. X., Li G. Y., Guo Y. Y., et al. (2017). Spectrum-effect relationships between fingerprints of Caulophyllum robustum maxim and inhabited pro-inflammation cytokine effects. Molecules 22:E1826. 10.3390/molecules22111826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan M. K., Ringoir D. D., Frirdich E., Svensson S. L., Wells D. H., Jarrell H., et al. (2008). Campylobacter jejuni biofilms up-regulated in the absence of the stringent response utilize a calcofluor white-reactive polysaccharide. J. Bacteriol. 190 1097–1107. 10.1128/jb.00516-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordalski S., Witek J., Smusz S., Rataj K., Bojarski A. J. (2015). Multiple conformational states in retrospective virtual screening - homology models vs. crystal structures: beta-2 adrenergic receptor case study. J. Cheminform. 7:13. 10.1186/s13321-015-0062-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus J., Wegrzyn G. (2001). Chloramphenicol-sensitive Escherichia coli strain expressing the chloramphenicol acetyltransferase (cat) gene. Antimicrob. Agents Chemother. 45 3610–3612. 10.1128/aac.45.12.3610-3612.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruess B. M., Verma K., Samanta P., Sule P., Kumar S., Wu J., et al. (2010). Environmental and genetic factors that contribute to Escherichia coli K-12 biofilm formation. Arch. Microbiol. 192 715–728. 10.1007/s00203-010-0599-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahni N., Yi S., Daniels K. J., Srikantha T., Pujol C., Soll D. R. (2009). Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 5:e1000601. 10.1371/journal.ppat.1000601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Liu Z., Liu C., Wu M., Su H., Ma X., et al. (2016). Spectrum-effect relationships between chemical fingerprints and antibacterial effects of lonicerae japonicae Flos and lonicerae Flos base on UPLC and microcalorimetry. Front. Pharmacol. 7:12. 10.3339/fphar.2016.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.-Y., Nam E.-H., Park S.-H., Hwang C.-Y. (2013). In vitro efficacy of the essential oil from Leptospermum scoparium (manuka) on antimicrobial susceptibility and biofilm formation in Staphylococcus pseudintermedius isolates from dogs. Vet. Dermatol. 24 404–408, e87. 10.1111/vde.12045 [DOI] [PubMed] [Google Scholar]
- Su G. Z., Cao Y., Li C., Yu X. L., Gao X. L., Tu P. F., et al. (2015). Phytochemical and pharmacological progress on the genus Syringa. Chem. Cent. J. 9:2 10.1186/s13065-015-0079-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulahong A. (2015). Molecular docking to understand the interaction between anti-tumor drug candidate gomisin j and cytochrome P450 3A4. Lat. Am. J. Pharm. 34 416–418. [Google Scholar]
- Venturini N., Barboni T., Curk F., Costa J., Paolini J. (2014). Volatile and flavonoid composition of the peel of Citrus medica L. var. Corsican fruit for quality assessment of its liqueur. Food Technol. Biotechnol. 52 403–410. 10.17113/ftb.52.04.14.3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang C., Gao L. F., Cai H., Zhou Y. H., Yang Y. B., et al. (2017). Rutin inhibits Streptococcus suis biofilm formation by affecting CPS biosynthesis. Front. Pharmacol. 8:379. 10.3389/fphar.2017.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li Y., Zhang Y., Feng G., Yang Z. X., Guan Q. X., et al. (2017). Multi-dimensional spectrum-effect relationship of the impact of chinese herbal formula lichong shengsui yin on ovarian cancer. Molecules 22:E979. 10.3390/molecules22060979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Yang Y. B., Zhao Y. L., Zhao H. H., Bai J. W., Chen J. Q., et al. (2016). Sub-MIC tylosin inhibits Streptococcus suis biofilm formation and results in differential protein expression. Front. Microbiol. 7:384. 10.3389/fmicb.2016.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yi L., Wu Z., Shao J., Liu G., Fan H., et al. (2012). Comparative proteomic analysis of Streptococcus suis biofilms and planktonic cells that identified biofilm infection-related immunogenic proteins. PLoS One 7:e33371. 10.1371/journal.pone.0033371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Wu Z. F., Zhu X. L., Lu C. P. (2011). Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet. Microbiol. 152 151–160. 10.1016/j.vetmic.2011.04.029 [DOI] [PubMed] [Google Scholar]
- Wong R. W. K., Haegg U., Samaranayake L., Yuen M. K. Z., Seneviratne C. J., Kao R. (2010). Antimicrobial activity of Chinese medicine herbs against common bacteria in oral biofilm. A pilot study. Int. J. Oral Maxillofac. Surg. 39 599–605. 10.1016/j.ijom.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. B., Wang S., Wang C., Huang Q. Y., Bai J. W., Chen J. Q., et al. (2015). Emodin affects biofilm formation and expression of virulence factors in Streptococcus suis ATCC700794. Arch. Microbiol. 197 1173–1180. 10.1007/s00203-015-1158-1154 [DOI] [PubMed] [Google Scholar]
- Yi T., Zhu L., Tang Y.-N., Zhang J.-Y., Liang Z.-T., Xu J., et al. (2014). An integrated strategy based on UPLC-DAD-QTOF-MS for metabolism and pharmacokinetic studies of herbal medicines: Tibetan ”Snow Lotus” herb (Saussurea laniceps), a case study. J. Ethnopharmacol. 153 701–713. 10.1016/j.jep.2014.03.031 [DOI] [PubMed] [Google Scholar]
- Zhang Y. J., Zhang X. (2007). Grey correlation analysis between strength of slag cement and particle fractions of slag powder. Cem. Concr. Compos. 29 498–504. 10.1016/j.cemconcomp.2007.02.004 [DOI] [Google Scholar]
- Zou P., Hong Y., Koh H. L. (2005). Chemical fingerprinting of Isatis indigotica root by RP-HPLC and hierarchical clustering analysis. J. Pharm. Biomed. Anal. 38 514–520. 10.1016/j.jpba.2005.01.022 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.