Abstract
Background
Diffuse large B cell lymphoma (DLBCL) contains heterogeneous subtypes with various molecular dysregulation at the gene, protein and microRNA levels. Compared with the GCB subtype, the non-germinal center B-like (non-GCB)/activated B cell-like (ABC) subtype exhibits frequent progression despite standard immunochemotherapy. We aimed to investigate the effects of miR-197 on the progression and chemosensitivity of DLBCL with respect to the GCB and non-GCB/ABC subtypes.
Methods
To screen distinctively expressed microRNAs, microRNA expression patterns were analyzed in 10 DLBCL cases by microarray chip assays. Using quantitative real-time polymerase chain reaction (qRT-PCR), associations between miR-197 expression levels and clinicopathologic variables were investigated in 51 DLBCL tissue samples. The effects of miR-197 on doxorubicin chemosensitivity were investigated using the OCI-Ly1 and SUDHL9 cell lines.
Results
MicroRNA expression profiling by hierarchical clustering revealed that miR-197 was one of the distinctively expressed microRNAs in DLBCL. Quantitative analysis using qRT-PCR revealed that miR-197 levels were not correlated with clinicopathologic variables, including the international prognostic index, but low miR-197 levels were significantly associated with lymphoma progression defined by refractoriness, relapse or death in the rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP)-treated subgroup (n = 43; p = 0.004). Among the three molecular groups, i.e., the GCB, non-GCB/miR-197low and non-GCB/miR-197high groups, progression was most frequently observed in the non-GCB/miR-197low group in the full cohort (p = 0.013) and the R-CHOP cohort (p = 0.008). In survival analysis, low miR-197 levels were independently predictive of shorter progression-free survival in the R-CHOP cohort (p = 0.031; HR = 27.9) and the non-GCB subgroup (p = 0.037; HR = 21.5) but not in the GCB subgroup. Using SUDHL9 (ABC type) and OCI-Ly1 (GCB type) cells, the effects of doxorubicin on reducing cell viability were enhanced by miR-197 transfection. In apoptosis assays, miR-197 transfection enhanced doxorubicin-induced apoptosis in SUDHL9 cells but not in OCI-Ly1 cells, suggesting a chemosensitizing effect of miR-197 in ABC DLBCL.
Conclusions
These results suggest the role of miR-197 as a biomarker with potential therapeutic implications.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1537-0) contains supplementary material, which is available to authorized users.
Keywords: microRNA, miR-197, Diffuse large B cell lymphoma, Chemosensitivity, Apoptosis
Background
Diffuse large B-cell lymphoma (DLBCL) is the most common type of malignant lymphoma, accounting for 30–40% of non-Hodgkin lymphoma cases [1]. DLBCL can be classified into two cell-of-origin (COO) subtypes, i.e., germinal center B cell (GCB) and activated B cell (ABC) types, which exhibit different gene expression profiles and clinical outcomes [2–6]. ABC DLBCL is recognized as a more aggressive subtype despite treatment with standard chemotherapy with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) [5]. Non-GCB DLBCL based on Hans and related algorithms using immunohistochemistry was inferior to GCB DLBCL regarding survival. In addition, non-GCB DLBCL is thought to represent ABC DLBCL with a defined gene expression signature [2, 3, 6] despite several inconsistent studies [7–9]. Although the clinical outcome of DLBCL can been distinctively improved with R-CHOP therapy, approximately 1/3 of DLBCL patients still experience relapse or refractoriness [10]. In this context, various studies have been performed to uncover and validate useful biomarkers to predict chemotherapeutic responses and clinical outcomes of DLBCL patients at the gene and protein levels as well as the post-transcriptional level of gene expression as modulated by microRNAs [11–13].
Doxorubicin is an anticancer drug in the anthracycline family and is used to treat various types of malignancies, including breast carcinoma, soft tissue sarcomas, acute leukemia and non-Hodgkin lymphomas [14–16]. Several complex mechanisms, including topoisomerase II inhibition, DNA intercalation, free radical generation, and the activation of several transcription factors, including cAMP responsive element binding protein 3-like 1 (CREB3L1), have been proposed to explain the antitumor activity of doxorubicin [17–19]. These biologic processes are regulated by many genes, proteins and microRNAs, and the response to this drug depends on the cellular context as it contains various regulatory molecules at the protein, gene and microRNA levels.
MicroRNAs are small non-coding RNAs composed of 20–24 nucleotides [20]. MicroRNAs are involved in many important biological processes, including cell proliferation, differentiation and apoptosis [21, 22], which might also be associated with critical oncogenic or tumor-suppressive pathways in neoplastic disease [23–25]. Therefore, the roles of microRNAs have been investigated in various types of solid and hematologic tumors, including DLBCL [26, 27], where distinct expression profiles were shown for GCB and ABC subtypes [28–30]. Several microRNAs are known to influence the sensitivity of tumor cells to anticancer drugs, consequently affecting the clinical outcomes of patients with hepatocellular carcinoma and chronic lymphocytic leukemia [31, 32]. MiR-451 is associated with doxorubicin resistance in breast cancer cells [33]. MiR-21 and miR-34a are regulators of chemosensitivity in DLBCL [34, 35]. Furthermore, miR-18a, miR-181a, miR-222, miR-199a and miR-497 predict overall and/or progression-free survival of DLBCL patients [36, 37].
Another important regulator of cancer-associated signaling in various solid and hematologic neoplasms is miR-197, which is involved in various pathways of cancer progression and chemoresistance depending on cell type and cellular context. MiR-197 regulates the response to 5-fluorouracil treatment in gastric and colorectal cancers via targeting mitogen-activated protein kinase 1 and thymidylate synthase, respectively [38, 39]. MiR-197 suppresses the p53-dependent apoptosis pathway in lung cancers [40] but induces apoptosis in myeloma cells via targeting Mcl-1 [41]. However, the role of miR-197 in DLBCL has not clearly been elucidated to date with respect to prognosis and chemoresistance.
In the present study, we aimed to investigate the effects of miR-197 in response to chemotherapy in DLBCL patients and to determine the possibility of miR-197 as an adjuvant therapeutic agent to enhance the effect of doxorubicin in DLBCL patients, especially those with the ABC subtype.
Methods
Patients and human tissue specimens
A total of 51 patients diagnosed with DLBCL at Seoul National University Hospital from 2006 to 2012 and Seoul National University Hospital from 2003 to 2012 were enrolled. The patients’ histologic slides and clinical medical records were retrospectively reviewed. According to the 2008 WHO classification of lymphoma, primary CNS lymphoma and mediastinal large B-cell lymphoma were excluded [42]. All DLBCL cases were divided into GCB and non-GCB subtypes using the Hans algorithm based on immunohistochemical findings [6]. The follow-up duration ranged from 0.5 to 64 months (median, 20.7 months). Twelve patients (24%) had progression and died at the time of analysis. Progression was defined as refractoriness, relapse or death. Progression-free survival (PFS) was defined as the time from first treatment to lymphoma progression, including death of any cause [43]. The institutional review board approved this study.
Cell lines and reagents
SUDHL9 and OCI-LY1 are human DLBCL cell lines. SUDHL9 cells were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) supplemented with 10% inactivated fetal bovine serum (WELGENE, Seoul, South Korea) and 1% antibiotic–antimycotic solution (WELGENE) in a humidified atmosphere with 5% CO2 in air at 37 °C. OCI-LY1 cells were cultured in Iscove’s Modified Dulbecco’s Medium (WELGENE) supplemented with 20% inactivated fetal bovine serum and 1% antibiotic–antimycotic solution. Doxorubicin (Cat no. D1515, Sigma-Aldrich, St. Louis, MO, USA) was purchased from Sigma-Aldrich.
RNA extraction from formalin-fixed tissues and cell lines
Total RNA was extracted from FFPE tissue samples using a RecoverAll™ Total Nucleic Acid Isolation for FFPE kit (Cat no. AM197, Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol and was stored at − 80 °C until the time of use after measuring the concentration with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA was extracted from DLBCL cell lines using TRI REAGENT (Cat no. TR118, Molecular Research Center, Cincinnati, OH, USA). MicroRNA was evaluated by qRT-PCR using Mir-X microRNA first strand synthesis and the SYBR qRT-PCR kit (Cat no. 638313, Clontech Laboratories, Mountain View, CA, USA). U6 served as an internal control.
Screening of microRNAs using a microarray chip test in DLBCL tissues
To screen distinctly expressed microRNAs in DLBCL, microRNA expression profiles in 10 formalin-fixed paraffin-embedded (FFPE) DLBCL tissue samples were investigated using a peptide nucleic acid (PNA)-based microRNA expression profiling kit (PANArray™, #PM-1001, version 1, Panagene, Daejeon, South Korea) containing 135 probes for cancer-related microRNAs according to the manufacturer’s instruction. U6 was included in the microarray slide as an internal control for normalization. The average value of the duplicate tests was used for expression profiling analysis. In this chip-based screening study, 73 microRNAs significantly expressed in DLBCL tissues were included in hierarchical clustering analysis of the agglomerative type (average linkage clustering). The DLBCL cases and microRNAs were reordered and clustered using the ‘heatmap.2’ function in the ‘gplots’ package in R using Euclidean measurements to obtain a distance matrix and the complete average agglomeration method [44].
Quantitative real-time polymerase chain reaction
Levels of hsa-miR-197 (cat no. 4427975, Applied Biosystems) were determined by qRT-PCR using TaqMan Universal PCR Master Mix and the TaqMan microRNA Reverse Transcription kit (Cat no. 4366596, Applied Biosystems), and U6 snRNA served as an internal control. The expression levels of the microRNAs were compared using 11 normal tonsil FFPE tissues as normal controls. The relative level of miR-197 in OSCC was calculated as 2−ΔΔCt, where ΔCt = Ct (miR-197) − Ct (U6) and ΔΔCt = ΔCt (tumor) − ΔCt (normal).
Cell culture and transfection of microRNA mimics
SUDHL9 and OCI-LY1 cells were seeded at a density of 3 × 105 cells/well in 12-well plates and were cultured in Opti-MEM (Gibco, Grand Island, NY, USA). For transfection, the transfection agent Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) and 50 and 100 nM microRNAs synthesized at GenePharma (Shanghai, China) were added to the cultured cells, and complementary media was added after 6 h.
Cell viability assay
SUDHL9 and OCI-LY1 cells were seeded at a density of 3 × 105 cells/well in 12-well plates. At 24 h post-transfection, various concentrations (0–100 nM for SUDHL9; 0–200 nM for OCI-LY1) of doxorubicin were added to the cells, resulting in a total volume of 1000 µl/well. At 48 h after doxorubicin treatment, 200 µl of treated cells were transfer to 96-well plates. Twenty microliters of EX-CYTOX (EX-3000, DOGEN Bio., Seoul, South Korea) agent was added to each well. Then, the cells were maintained in an incubator with 5% CO2 at 37 °C for 2–3 h. The absorbance at 450 nm, which is indicative of cell viability, was detected. All reactions were analyzed in triplicate.
Apoptosis assay
SUDHL9 and OCI-LY1 cells were treated with the same transfection conditions. Twenty-four hours post-transfection, SUDHL9 cells were treated with 25 and 50 nM doxorubicin for 48 h, and OCI-LY1 cells were treated with 50, 100 and 200 nM doxorubicin for 72 h. For analysis of apoptosis, cells were stained with 3 µl of Annexin V and 1.5 µl of PI for 10 min at RT in the dark using the FITC Annexin V Apoptosis Detection Kit I (556547, BD Biosciences, San Jose, CA, USA). All reactions were analyzed in triplicate using BD flow cytometry (BD FACScalibur, BD Biosciences).
Statistical analysis
The microRNA expression data obtained from microarray chip assays for ten cases of DLBCL were analyzed by hierarchical clustering with dendrograms using R package 3.1.2 (gplots). Dot plots for comparison of microRNA expression levels between subgroups were also generated by R package 3.1.2 (ggplot2 and beeswarm). Other statistical analyses were performed using SPSS 21.0 (IBM, Armonk, NY, USA). All experiments were independently performed in triplicate. Independent sample t-tests were used for comparison of microRNA expression levels. Survival data were analyzed using Kaplan–Meier analysis with log-rank tests and Cox proportional hazard models. p-values less than 0.05 were considered statistically significant. MiR-197 expression was categorized as high or low expression based on the cutoff point exhibiting maximum χ2 (minimum p-value) as a prognostic factor [45]. This method employs a systematic search of almost all observed values as the candidate cutoff point and selects the value associated with a maximum χ2 (or minimum p value) as the final cutoff point [45].
Results
Screening of distinctively expressed microRNAs and identification of miR-197 in DLBCL
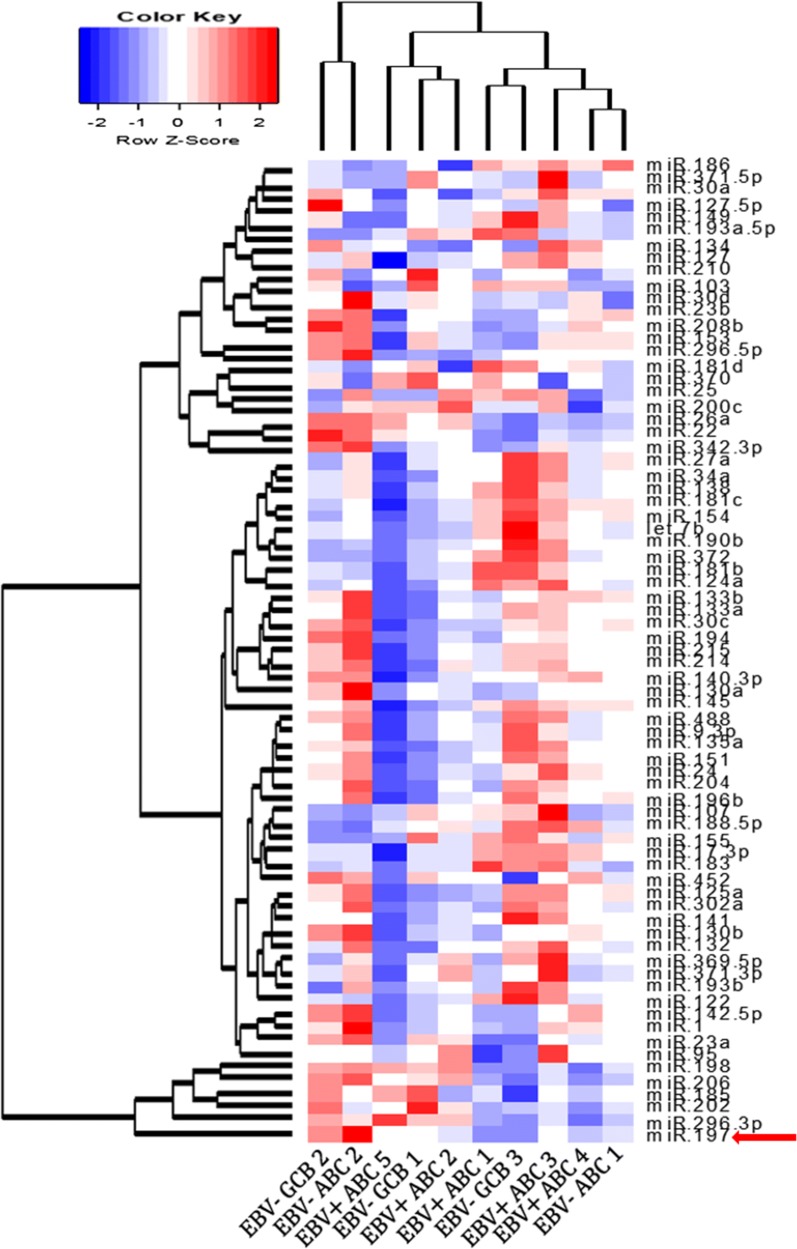
By analyzing expression profiling using the cancer-related microRNA microarray platform, six microRNAs (miR-197, miR-296-3p, miR-202, miR-185, miR-206 and miR-198) were recognized to form a distinctive cluster in DLBCLs, as shown in the heatmap with dendrograms generated using a hierarchical clustering algorithm (Fig. 1 and Additional file 1: Figure S1). Among the six microRNAs, miR-197 exhibited the most distinctive pattern in the cluster, and miR-197 expression levels exhibited wide ranges among the various subtypes of DLBCLs. Therefore, we focused on the quantitative analysis of miR-197 to investigate its clinicopathologic association in DLBCLs.
Fig. 1.
Heatmap with dendrograms generated by hierarchical clustering in DLBCLs. Based on a hierarchical clustering algorithm, six microRNAs (miR-197, miR-296-3p, miR-202, miR-185, miR-206 and miR-198) were distinctively clustered irrespective of DLBCL subtypes. MiR-197 is marked with a red arrow
Clinicopathologic features of diffuse large B cell lymphoma patients
The clinicopathologic features of 51 cases of DLBCL are presented in Table 1. Among the 51 cases of DLBCL, 22% (11/51) were EBV positive. Old age (≥ 60 years), male patients, extranodal cases and low international prognostic index (IPI) were common features, accounting for approximately 60% of cases. Based on the Hans algorithm, 73% (37/51) of cases were classified as the non-GCB type, which included all 11 cases of EBV-positive DLBCL. R-CHOP therapy was administered to 84% (43/51) of patients.
Table 1.
Clinicopathologic characteristics of diffuse large B cell lymphoma patients
| Variables | Full cohort (n = 51) | R-CHOP cohort (n = 43) |
|---|---|---|
| Number of cases (%) | Number of cases (%) | |
| Age, years | ||
| Mean (range) | 59.6 (21–85) | |
| < 60 | 21 (41%) | 17 (40%) |
| ≥ 60 | 30 (59%) | 26 (60%) |
| Sex | ||
| Male | 30 (59%) | 24 (56%) |
| Female | 21 (41%) | 19 (44%) |
| Primary site | ||
| Nodal | 18 (35%) | 14 (33%) |
| Extranodal | 33 (65%) | 29 (67%) |
| Ann Arbor stage | ||
| I, II | 27 (53%) | 23 (54%) |
| III, IV | 24 (37%) | 20 (46%) |
| IPI groupa | ||
| Low (0–2) | 30 (60%) | 27 (63%) |
| High (3–5) | 20 (40%) | 16 (37%) |
| B symptomsa | ||
| Absent | 31 (62%) | 29 (67%) |
| Present | 19 (38%) | 14 (33%) |
| ECOG PSa | ||
| 0, 1 | 40 (82%) | 37 (86%) |
| ≥ 2 | 9 (18%) | 6 (14%) |
| LDHa | ||
| Normal | 21 (42%) | 19 (44%) |
| Elevated | 29 (58%) | 24 (56%) |
| BM involvementa | ||
| Absent | 42 (91%) | 38 (93%) |
| Present | 4 (9%) | 3 (7%) |
| Number of extranodal sitesa | ||
| 0, 1 | 40 (80%) | 33 (77%) |
| ≥ 2 | 10 (20%) | 10 (23%) |
| EBER | ||
| Negative | 40 (78%) | 36 (84%) |
| Positive | 11 (22%) | 7 (16%) |
| Hans classification | ||
| GCB | 14 (27%) | 12 (28%) |
| Non-GCB | 37 (73%) | 31 (72%) |
| Treatment | ||
| Rituximab + CHOP | 43 (84%) | 43 (100%) |
| Rituximab + others | 1 (2%) | 0 (0%) |
| Surgery only | 2 (4%) | 0 (0%) |
| No Tx | 5 (10%) | 0 (0%) |
IPI international prognostic index, ECOG PS Eastern Cooperative Oncology Group Performance Status, LDH lactate dehydrogenase, BM bone marrow, EBER EBV-encoded RNA, GCB germinal center B-like, CHOP cyclophosphamide, doxorubicin, vincristine, and prednisone
aThese variables exclude missing values
MiR-197 expression levels according to different clinicopathologic groups
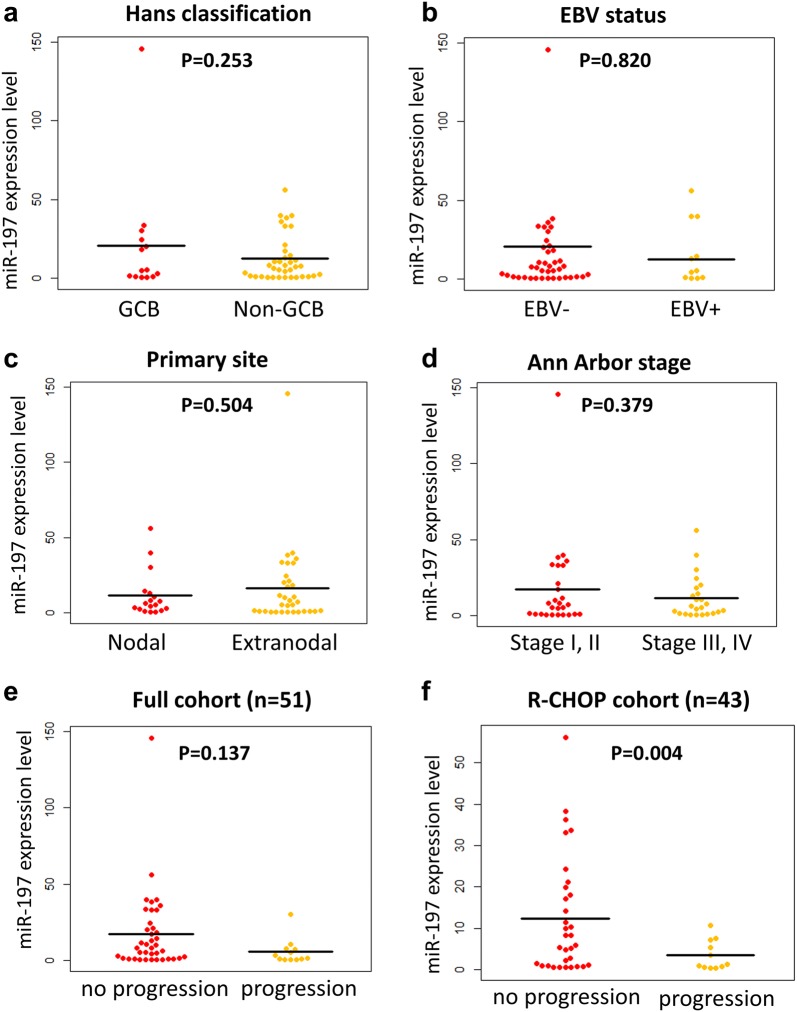
As shown in Fig. 2a–d and Table 2, miR-197 expression levels were compared between various subgroups and clinicopathologic factors of DLBCL, i.e., EBV-positive vs. EBV-negative cases; GCB vs. non-GCB/ABC type; and nodal vs. extranodal cases. Based on comparisons of mean values using independent t-tests, miR-197 expression levels were not significantly associated with EBV status, nodal/extranodal, stages (I, II vs. III, IV) and IPI (0–2 vs. 3–5). The non-GCB subtype tended to have low miR-197 expression levels, but the result did not achieve statistical significance (p = 0.253).
Fig. 2.
Relative miR-197 expression level based on clinicopathologic variables and progression status. Relative miR-197 expression levels did not differ based on Hans classification (a), EBV infection (b), primary site (c) or Ann Arbor stage (d) in the full cohort of diffuse large B-cell lymphoma (n = 51). MiR-197 levels tended to be reduced in the progression group compared with the no progression group in the full cohort (n = 51) (e). In the R-CHOP cohort (n = 43), miR-197 was significantly reduced in the progression group (f)
Table 2.
Comparison of miR-197 expression levels according to clinicopathologic characteristics in diffuse large B cell lymphoma patients
| Variables | Full cohort (n = 51) | R-CHOP cohort (n = 43) | ||
|---|---|---|---|---|
| Mean ± SD | p value | Mean ± SD | p value | |
| Age, years | 0.415 | 0.357 | ||
| < 60 | 18.2 ± 32.6 | 7.8 ± 12.0 | ||
| ≥ 60 | 12.0 ± 13.7 | 11.6 ± 13.4 | ||
| Sex | 0.603 | 0.353 | ||
| Male | 16.0 ± 27.7 | 8.5 ± 10.4 | ||
| Female | 12.5 ± 15.4 | 12.2 ± 15.4 | ||
| Primary site | 0.504 | 0.595 | ||
| Nodal | 11.5 ± 15.3 | 8.6 ± 14.3 | ||
| Extranodal | 16.2 ± 26.8 | 10.8 ± 12.3 | ||
| Ann Arbor stage | 0.379 | 0.731 | ||
| I, II | 17.3 ± 29.2 | 10.7 ± 12.8 | ||
| III, IV | 11.5 ± 14.1 | 9.4 ± 13.2 | ||
| IPI groupa | 0.664 | 0.985 | ||
| Low (0–2) | 11.2 ± 13.0 | 10.1 ± 12.3 | ||
| High (3–5) | 13.0 ± 15.6 | 10.1 ± 14.1 | ||
| B symptomsa | 0.737 | |||
| Absent | 15.7 ± 27.4 | 10.4 ± 12.2 | ||
| Present | 13.4 ± 15.6 | 9.5 ± 14.4 | ||
| ECOG PSa | 0.886 | 0.310 | ||
| 0, 1 | 12.0 ± 13.8 | 10.9 ± 13.5 | ||
| ≥ 2 | 12.8 ± 16.0 | 5.1 ± 5.9 | ||
| LDHa | 0.466 | 0.785 | ||
| Normal | 10.2 ± 13.2 | 9.5 ± 12.6 | ||
| Elevated | 13.2 ± 14.6 | 10.6 ± 13.3 | ||
| BM involvementa | 0.561 | 0.452 | ||
| Absent | 11.0 ± 12.5 | 9.1 ± 11.2 | ||
| Present | 19.2 ± 24.8 | 24.1 ± 27.9 | ||
| Number of extranodal sitesa | 0.182 | 0.582 | ||
| 0, 1 | 12.9 ± 15.0 | 10.7 ± 14.0 | ||
| ≥ 2 | 8.1 ± 8.0 | 8.1 ± 8.0 | ||
| EBER | 0.820 | 0.804 | ||
| Negative | 14.1 ± 24.4 | 9.9 ± 11.2 | ||
| Positive | 16.0 ± 19.8 | 11.2 ± 20.4 | ||
| Hans classification | 0.253 | 0.833 | ||
| GCB | 20.7 ± 38.0 | 9.4 ± 11.5 | ||
| Non-GCB | 12.2 ± 14.6 | 10.4 ± 13.5 | ||
IPI international prognostic index, ECOG PS Eastern Cooperative Oncology Group Performance Status, LDH lactate dehydrogenase, BM bone marrow, EBER EBV-encoded RNA, GCB germinal center B-like
aThese variables exclude missing values
Differential expression levels of miR-197 based on progression status
MiR-197 expression levels were compared between the patients who had progression, i.e., refractoriness, relapse, or death, and those who did not exhibit progression during the follow-up period (Fig. 2e, f). Patients who progressed exhibited significantly low miR-197 levels in the R-CHOP-treated cohort (p = 0.004; n = 43), whereas the difference was not significant in the full cohort (p = 0.137; n = 51).
Associations among progression status, GCB/non-GCB subtype and miR-197 expression
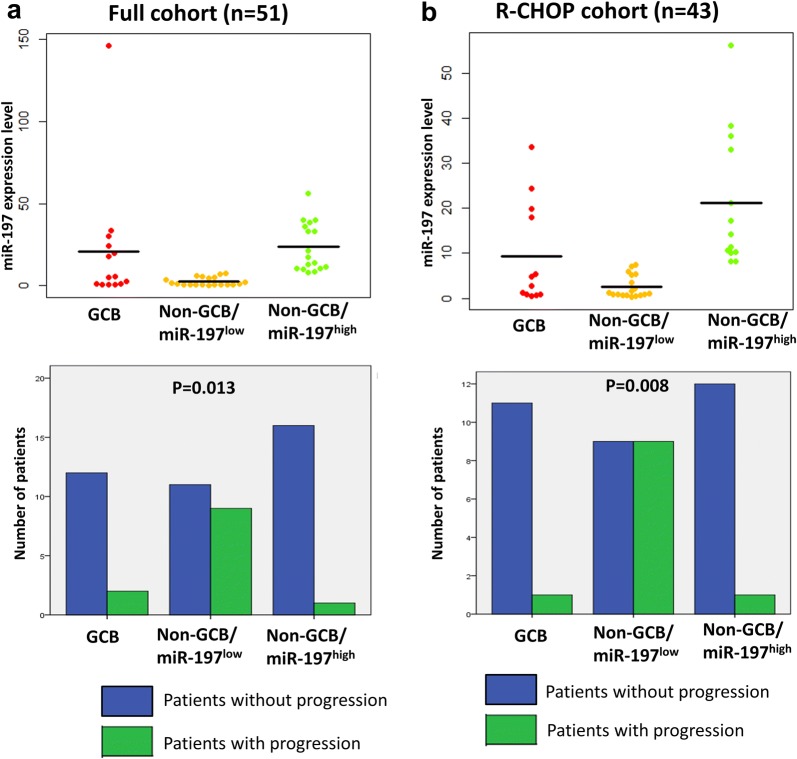
Considering the significant effects of miR-197 on progression within the non-GCB subgroup, we analyzed the frequencies of progression among the three molecular groups, i.e., GCB, non-GCB/miR-197low and non-GCB/miR-197high groups. As shown in Fig. 3, progression was most frequently noted in the non-GCB/miR-197low group in the full cohort (p = 0.013) and the R-CHOP cohort (p = 0.008).
Fig. 3.
Comparison of relative miR-197 expression levels and frequency of patients with progression in the GCB, non-GCB/miR-197low and non-GCB/miR-197high subgroups of diffuse large B-cell lymphoma. Compared with the GCB and non-GCB/miR-197high subgroups exhibiting similar levels of miR-197 expression, the non-GCB/miR-197low subgroup exhibited low miR-197 levels and high progression rates in full cohort (a) and R-CHOP cohort (b)
Survival analysis of progression-free survival based on miR-197 expression levels
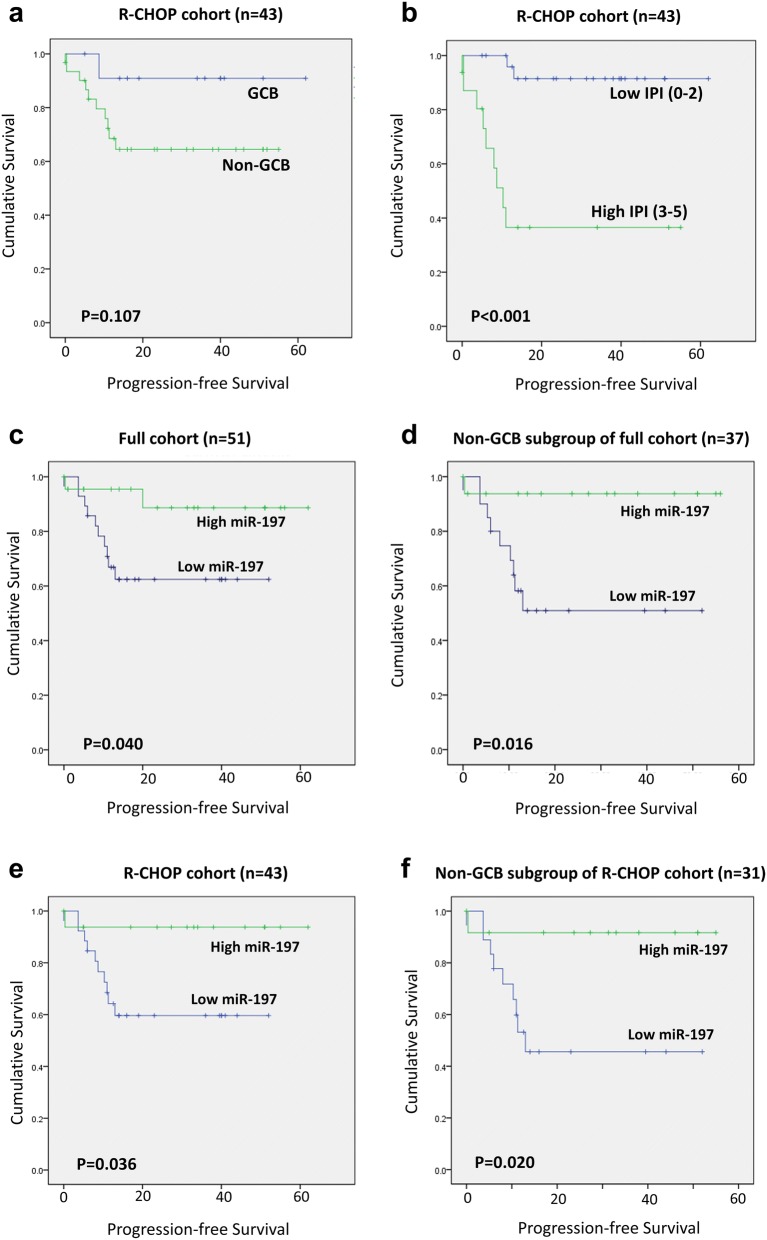
Given the close relationship between progression status and miR-197 expression level, the effects of miR-197 and various clinicopathologic variables on PFS were analyzed using the Kaplan–Meier method with log-rank tests. As shown in Fig. 4, the GCB type tended to have a good prognosis without statistical significance (p = 0.107; Fig. 4a), whereas low IPI was significantly associated with longer PFS (p < 0.001; Fig. 4b). The miR-197low group exhibited reduced PFS compared with the miR-197high group in the full cohort (n = 51; n = 0.040; Fig. 4c) and its non-GCB subgroup (n = 37; p = 0.016; Fig. 4d) as well as the R-CHOP cohort (n = 43; p = 0.036; Fig. 4e) and its non-GCB subgroup (n = 31; p = 0.020; Fig. 4f). In contrast, the prognostic significance of miR-197 was not evident in the GCB subgroup of the full cohort or R-CHOP cohort (p > 0.05).
Fig. 4.
Survival curves for progression-free survival in diffuse large B-cell lymphoma. The non-germinal center B-like (non-GCB) subtype tended to exhibit shorter progression-free survival (PFS) compared with the GCB subtype (a), whereas a high international prognostic index (IPI) was a significant predictor of shorter PFS (b) in the R-CHOP cohort (n = 43). Low miR-197 levels were also associated with shorter PFS in the full cohort (n = 51) (c) and its non-GCB subgroup (n = 37) (d) as well as the R-CHOP cohort (n = 43) (e) and its non-GCB subgroup (n = 31) (f)
Progression-free survival analysis based on miR-197 expression levels and clinicopathologic prognostic factors in the R-CHOP cohort
Table 3 summarizes the prognostic factors predicting PFS using univariate and multivariate analysis in the R-CHOP cohort (n = 43) and ABC subgroup (n = 31). In the R-CHOP cohort, miR-197 was associated with PFS in univariate analysis. Other prognostic factors included IPI, B symptoms, bone marrow involvement, stage, performance status, lactate dehydrogenase (LDH), and extranodal site number. Multivariate analysis revealed that IPI (p = 0.009; HR = 10.0), miR-197 (p = 0.031; HR = 27.9), and bone marrow involvement (p = 0.038; HR = 15.4) predicted PFS. In the non-GCB subgroup, univariate analysis revealed similar prognostic factors, including miR-197 and other clinicopathologic factors. Multivariate analysis incorporating miR-197, IPI and bone marrow involvement revealed that miR-197 (p = 0.037; HR = 21.5) and bone marrow involvement (p = 0.046; HR = 13.5) were significant prognostic factors.
Table 3.
Univariate and multivariate survival analysis of clinicopathologic variables and miR-197 expression in diffuse large B cell lymphoma patients
| Clinicopathologic variables | R-CHOP cohort, n = 43 | ABC R-CHOP cohort, n = 31 | ||||
|---|---|---|---|---|---|---|
| UA | MA | UA | MA | |||
| p | p | HR [95% CI] | p | p | HR [95% CI] | |
| miR-197 | ||||||
| Low vs. high | 0.036 | 0.031 | 27.9 [1.4–569.0] | 0.025 | 0.037 | 21.5 [1.2–382.6] |
| IPI | ||||||
| High (3–5) vs. low (0–2) | < 0.001 | 0.009 | 10.0 [1.8–56.1] | 0.001 | 0.055 | 5.1 [1.0–26.6] |
| B symptoms | ||||||
| Presence vs. absence | 0.024 | 0.844 | 1.2 [0.3–4.8] | 0.059 | ||
| Bone marrow involvement | ||||||
| Presence vs. absence | 0.006 | 0.038 | 15.4 [1.2–203.3] | 0.029 | 0.046 | 13.5 [1.1–173.1] |
| Stagea | ||||||
| I–II vs. III–IV | 0.004 | 0.012 | ||||
| Performance statusa | ||||||
| ≥ 2 vs. 0–1 | < 0.001 | < 0.001 | ||||
| LDHa | ||||||
| Elevated vs. normal | 0.005 | 0.005 | ||||
| Number of extranodal sitesa | ||||||
| ≥ 2 vs. 0–1 | < 0.001 | 0.002 | ||||
UA univariate analysis, MA multivariate analysis, 95% CI 95% confidence interval, IPI international prognostic index, LDH lactate dehydrogenase
aThese variables were not included in multivariate analysis due to their close relationship with IPI
Cell viability assay after doxorubicin and miR-197 transfection treatment in DLBCL cell lines
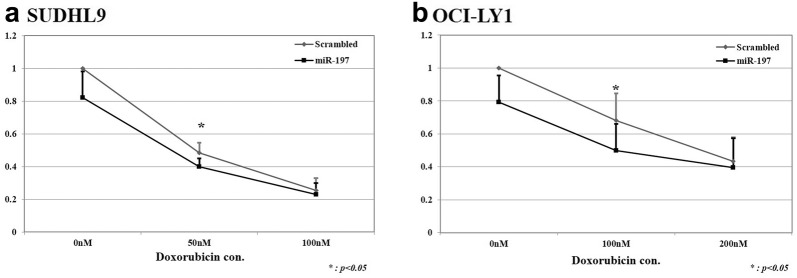
Given the results from clinical samples demonstrating that miR-197 influenced the progression status and PFS, we hypothesized that miR-197 may be associated with chemoresistance in DLBCL tumor cells. Next, we selected two cell lines with low miR-197 expression to investigate the role of miR-197 in DLBCL cell lines treated with doxorubicin, a key tumoricidal component of the R-CHOP chemotherapeutic regimen. SUDHL9, an ABC type DLBCL cell line, and OCI-Ly1, a GCB type DLBCL cell line, cells were treated with doxorubicin with or without miR-197 mimic transfection. Upon transfection with the miR-197 mimic, cell viability significantly decreased after doxorubicin treatment in SUDHL9 and OCI-Ly1 cells compared with that in the scrambled control group (Fig. 5).
Fig. 5.
Cell viability assay results for doxorubicin treatment and miR-197 transfection in diffuse large B-cell lymphoma cell lines. Upon treatment with a series of doxorubicin concentrations in diffuse large B-cell lymphoma cells of non-germinal center/activated B cell-like (non-GCB/ABC; SUDHL9) phenotype and GCB origin (OCI-LY1), cell viability was significantly reduced in the miR-197 transfection group compared with that in the scrambled control group. SUDHL9, an ABC DLBCL cell line, exhibited significant miR-197-transfection-induced reduction in cell viability at lower concentrations of doxorubicin (a) compared with OCI-Ly1, a GCB DLBCL cell line (b)
Apoptosis assay
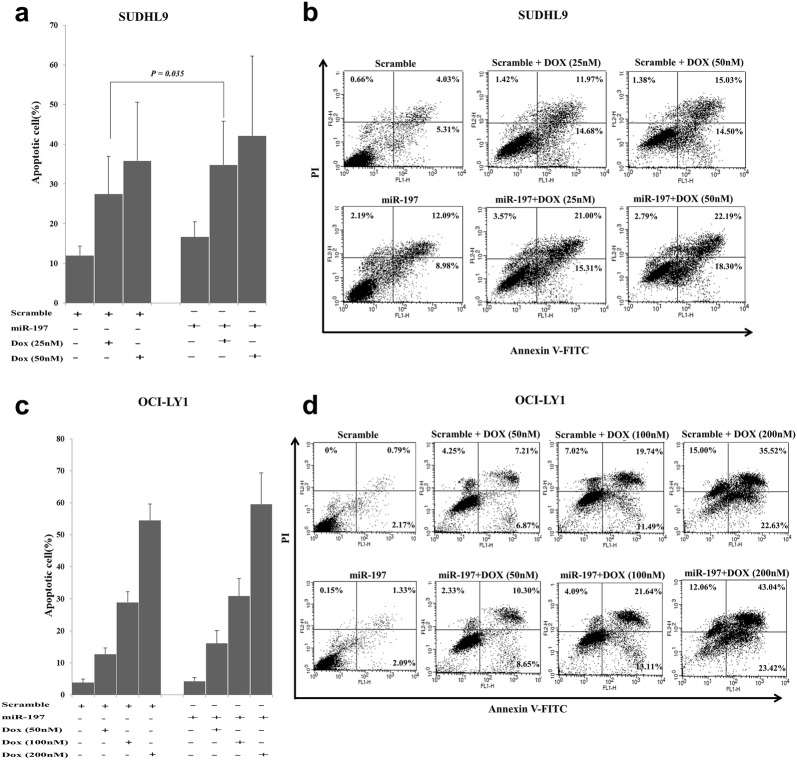
In the apoptosis assay with Annexin V and propidium iodide (PI) staining using flow cytometry, the effects of miR-197 differed between SUDHL9 and OCI-Ly1 cells (Fig. 6). In SUDHL9 cells, miR-197 mimic transfection produced a significant increase in the proportion of apoptotic cells in the 25 nM doxorubicin treatment group, up to the similar level of the 50 nM doxorubicin treatment control group, suggesting a chemosensitizing effect of miR-197 (Fig. 6a, b). In contrast, OCI-Ly1 cells did not exhibit a significant difference in the proportion of apoptotic cells upon transfection with miR-197 mimic in each concentration group of doxorubicin treatment (Fig. 6c, d). Therefore, miR-197 might have an important role in doxorubicin chemosensitivity by enhancing doxorubicin-induced apoptosis in SUDHL9, an ABC DLBCL cells. This finding is consistent with the results from patient samples from the R-CHOP cohort and the ABC subgroup.
Fig. 6.
Apoptosis assay results for doxorubicin treatment and miR-197 transfection in diffuse large B-cell lymphoma cell lines. Measurement of apoptotic cell proportions upon transfection of miR-197 in SUDHL9, an activated B cell-like diffuse large B-cell lymphoma (ABC DLBCL) cell line, revealed the induction of a significant chemosensitizing effect to 25 nM doxorubicin treatment as shown in the bar graph (a) and representative flow cytometry plot (b), whereas no significant difference was observed in OCI-LY1, a germinal center B-like DLBCL cell line, as shown in the bar graph (c) and representative flow cytometry plot (d)
Discussion
Treatment outcome of DLBCL has been considerably improved given the development of combination chemotherapy and the introduction of rituximab. However, approximately 1/3 of patients with DLBCL still experience relapse or refractoriness after standard therapy, eventually exhibiting a very poor outcome [10]. Among the numerous clinical and molecular subtypes of DLBCL, the ABC subtype exhibits a high rate of relapse or refractoriness, and targeting strategies focusing on the ABC subtype are being developed while its clinical utility still remains to be validated [46, 47]. Considering that microRNAs interfere with multiple targets involved in cell survival and drug resistance, we focused on the role of miR-197 in DLBCL and investigated whether it could determine the responsiveness of standard R-CHOP therapy and enhance the sensitivity of doxorubicin in DLBCL cells. In the present study, we observed that (1) miR-197 expression levels were significantly low in the DLBCL progression group, (2) low miR-197 levels were associated with shorter PFS in the R-CHOP cohort and ABC subgroup, and (3) miR-197 enhanced doxorubicin sensitivity in SUDHL9 cells of the ABC subtype.
MiR-197 is involved in apoptosis in solid tumors and hematologic neoplasms. In lung cancers, miR-197 suppresses apoptosis in a p53-dependent manner, playing an oncogenic role [40]. In contrast, a different role of miR-197 in apoptosis is noted in hematologic cells. In myeloma cells, miR-197 and miR-137 induce apoptosis by targeting Mcl-1 [41]. The researchers focused on frequent loss of the 1p12-21 region containing miR-137/miR-197 genes and demonstrated reduction of Mcl-1 protein and induction of apoptosis upon transfection of miR-137/miR-197 into myeloma cells as well as in vivo tumor regression by intratumoral injection of miR-137/miR-197 in a xenograft murine model of myeloma. The inhibiting effect of miR-197 to Mcl-1 may also be at least partly effective in ABC DLBCL because gain or amplification of the Mcl-1 gene was noted in 26% of ABC DLBCL [48]. ABC DLBCLs are thought to be derived from B cells at a plasmablastic stage frequently expressing genes of mature plasma cells [49, 50] and exhibit CBM complex (CARD11, BCL10, and MALT1)-driven constitutive activation of NF-κB signaling [51, 52], which is a key oncogenic signaling pathway that activates Mcl-1 in multiple myeloma [53].
Another important oncogenic signaling pathway in ABC DLBCL is the cytokine signaling/Janus kinase–signal transducer and activator of transcription (JAK–STAT) pathway [49]. A study using ABC DLBCL cells revealed that aberrant expression of IL-22 receptor 1 and IL-22 stimulation increased STAT3 and ERK1/2 activation, suggesting the important pathogenic roles of the IL-22/IL-22R1/STAT3 pathway in ABC DLBCL [54]. Remarkably, miR-197 reduces STAT3 signaling via miR-197/IL-22/STAT3 and miR-197/IL-6/STAT3 pathways in human keratinocytes and hepatocellular carcinomas [55, 56]. In addition, MyD88 mutations occur in > 30% of ABC DLBCL cases and activate both NF-κB and JAK–STAT signaling pathways [57], both of which participate in anti-apoptosis, cell survival and chemoresistance [49, 58, 59].
DLBCL exhibits heterogeneous clinical outcomes and molecular alterations [49]. The wide distribution of miR-197 expression levels in our data may reflect the heterogeneous molecular nature of DLBCL and its ABC subtype. Considering that microRNAs constitute complex multiple network of signaling pathways [60], relative expression levels of miR-197 may indicate inhibition of NF-κB/Mcl-1 and STAT3 signaling in ABC DLBCL, suggesting the possible role of miR-197 as a biomarker of progression in DLBCL, especially in the ABC subtype.
DLBCL heterogeneity was also demonstrated in hierarchical clustering analysis of microRNA chip-based expression profiling, where the cases were not clustered based on disease type or EBV status. This finding may partly be associated with the general cancer-related but not lymphoma-specific properties of the microRNA panel and the relatively small number of cases analyzed. Based on this type of screening, we observed that a miR-197 was distinctively identified and was associated with lymphoma progression and chemoresistance, which could be a cancer-related property, as demonstrated in the clinicopathologic analysis with clinical samples and the in vitro assays.
In vitro studies using the SUDHL9 cell line of the ABC subtype revealed that the effect of low-level (25 nM) doxorubicin treatment was approximately equivalent to treatment with twice (50 nM) the concentration. This effect might suggest the possibility of miR-197 as a chemosensitizing adjuvant agent to reduce doxorubicin concentrations, which might provide the same tumor-killing effect and reduce the cardiac toxicity associated with doxorubicin. This strategy to supplement miR-197 requires further clarification with pre-clinical and clinical studies.
In our in vitro investigation, we focused on the role of miR-197 in doxorubicin sensitivity because doxorubicin is an essential component of chemotherapeutic regimens that kill aggressive B lymphoma cells, including the standard R-CHOP regimen as well as R-EPOCH (rituximab, etoposide, prednisone, vincristine, cyclophosphamide and doxorubicin) and hyper-CVAD (course A: cyclophosphamide, vincristine, doxorubicin, and dexamethasone; course B: methotrexate and cytarabine) [61]. Moreover, sufficient doses of doxorubicin are critical for clinical outcome in DLBCL patients [62]. It still remains to be clarified further whether miR-197 may also influence the effects on sensitivity of other drugs.
Conclusions
Taken together, the results in our study suggests the clinicopathologic implication of miR-197 in DLBCL. Low miR-197 expression is associated with frequent progression and reduced PFS in DLBCL after standard R-CHOP therapy, especially in the ABC subtype. In vitro studies reveal a chemosensitizing effect of miR-197 in SUDHL9 cells of the ABC subtype treated with doxorubicin. These findings suggest a possible role of miR-197 as a useful biomarker to predict progression and as a chemosensitizer to enhance therapeutic efficacy, providing insight into effective therapeutic strategies with detailed risk assessment in DLBCL.
Additional file
Additional file 1: Fiugre S1. Hierarchical clustering of diffuse large B cell lymphoma cases and microRNA expression profiling with complete agglomerative (a), single agglomerative (b), Ward method (c), and divisive method (d).
Authors’ contributions
JMY, YKJ and JP conceived and designed the experiments. JMY and JYJ performed the experiments. JMY and JHP wrote the manuscript. All authors edited the final manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The Institutional Review Board of Seoul National University Bundang Hospital (IRB of SNUBH) approved this study, and informed consent was waived by IRB of SNUBH because this study used archived materials with retrospective design.
Funding
This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) of Korea funded by the Ministry of Education (NRF-2016R1D1A1B03935507) and a Grant (03-2011-015) from the Seoul National University Bundang Hospital Research Fund.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DLBCL
diffuse large B cell lymphoma
- GCB
germinal center B-like
- ABC
activated B cell-like
- R-CHOP
rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone
- IPI
international prognostic index
- FFPE
formalin-fixed paraffin-embedded
- LDH
lactate dehydrogenase
- CBM complex
CARD11, BCL10, and MALT1 complex
- JAK–STAT
Janus kinase–signal transducer and activator of transcription
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1537-0) contains supplementary material, which is available to authorized users.
Contributor Information
Jeong Mi Yang, Email: didwjdal1210@nate.com.
Ji-Young Jang, Email: jjy93@hanmail.net.
Yoon Kyung Jeon, Email: ykjeon@snu.ac.kr.
Jin Ho Paik, Phone: +82-31-787-7717, Email: paikjh@snu.ac.kr.
References
- 1.Scott DW. Cell-of-origin in diffuse large B-cell lymphoma: are the assays ready for the clinic? Am Soc Clin Oncol Educ Book. 2015;35:e458–e466. doi: 10.14694/EdBook_AM.2015.35.e458. [DOI] [PubMed] [Google Scholar]
- 2.Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, Braziel RM, Geng H, Iqbal J, Lenz G, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, Ott G, Rosenwald A, Braziel RM, Campo E, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–207. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muris JJ, Meijer CJ, Vos W, van Krieken JH, Jiwa NM, Ossenkoppele GJ, Oudejans JJ. Immunohistochemical profiling based on Bcl-2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B cell lymphoma. J Pathol. 2006;208:714–723. doi: 10.1002/path.1924. [DOI] [PubMed] [Google Scholar]
- 5.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 6.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 7.Paik JH, Go H, Nam SJ, Kim TM, Heo DS, Kim CW, Jeon YK. Clinicopathologic implication of A20/TNFAIP3 deletion in diffuse large B-cell lymphoma: an analysis according to immunohistochemical subgroups and rituximab treatment. Leuk Lymphoma. 2013;54:1934–1941. doi: 10.3109/10428194.2012.762511. [DOI] [PubMed] [Google Scholar]
- 8.Colomo L, Lopez-Guillermo A, Perales M, Rives S, Martinez A, Bosch F, Colomer D, Falini B, Montserrat E, Campo E. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood. 2003;101:78–84. doi: 10.1182/blood-2002-04-1286. [DOI] [PubMed] [Google Scholar]
- 9.Gutierrez-Garcia G, Cardesa-Salzmann T, Climent F, Gonzalez-Barca E, Mercadal S, Mate JL, Sancho JM, Arenillas L, Serrano S, Escoda L, et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood. 2011;117:4836–4843. doi: 10.1182/blood-2010-12-322362. [DOI] [PubMed] [Google Scholar]
- 10.Sehn LH, Donaldson J, Chhanabhai M, Fitzgerald C, Gill K, Klasa R, MacPherson N, O’Reilly S, Spinelli JJ, Sutherland J, et al. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B-cell lymphoma in British Columbia. J Clin Oncol. 2005;23:5027–5033. doi: 10.1200/JCO.2005.09.137. [DOI] [PubMed] [Google Scholar]
- 11.Klapper W, Stoecklein H, Zeynalova S, Ott G, Kosari F, Rosenwald A, Loeffler M, Trumper L, Pfreundschuh M, Siebert R. Structural aberrations affecting the MYC locus indicate a poor prognosis independent of clinical risk factors in diffuse large B-cell lymphomas treated within randomized trials of the German high-grade Non-Hodgkin’s lymphoma study group (DSHNHL) Leukemia. 2008;22:2226–2229. doi: 10.1038/leu.2008.230. [DOI] [PubMed] [Google Scholar]
- 12.Savage KJ, Johnson NA, Ben-Neriah S, Connors JM, Sehn LH, Farinha P, Horsman DE, Gascoyne RD. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–3537. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 13.Go H, Jang JY, Kim PJ, Kim YG, Nam SJ, Paik JH, Kim TM, Heo DS, Kim CW, Jeon YK. MicroRNA-21 plays an oncogenic role by targeting FOXO1 and activating the PI3K/AKT pathway in diffuse large B-cell lymphoma. Oncotarget. 2015;6:15035–15049. doi: 10.18632/oncotarget.3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, et al. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 15.D’Adamo DR, Anderson SE, Albritton K, Yamada J, Riedel E, Scheu K, Schwartz GK, Chen H, Maki RG. Phase II study of doxorubicin and bevacizumab for patients with metastatic soft-tissue sarcomas. J Clin Oncol. 2005;23:7135–7142. doi: 10.1200/JCO.2005.16.139. [DOI] [PubMed] [Google Scholar]
- 16.Guo B, Zhu HL, Li SX, Lu XC, Fan H. Individualized liposomal doxorubicin-based treatment in elderly patients with non-Hodgkin’s lymphoma. Onkologie. 2011;34:184–188. doi: 10.1159/000327008. [DOI] [PubMed] [Google Scholar]
- 17.Denard B, Lee C, Ye J. Doxorubicin blocks proliferation of cancer cells through proteolytic activation of CREB3L1. Elife. 2012;1:e00090. doi: 10.7554/eLife.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meredith AM, Dass CR. Increasing role of the cancer chemotherapeutic doxorubicin in cellular metabolism. J Pharm Pharmacol. 2016;68:729–741. doi: 10.1111/jphp.12539. [DOI] [PubMed] [Google Scholar]
- 19.Agudelo D, Bourassa P, Berube G, Tajmir-Riahi HA. Intercalation of antitumor drug doxorubicin and its analogue by DNA duplex: structural features and biological implications. Int J Biol Macromol. 2014;66:144–150. doi: 10.1016/j.ijbiomac.2014.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 21.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 22.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 25.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 26.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 28.Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood J, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 29.Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, Thiere M, Loeffler M, Klapper W, Pfreundschuh M, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 30.Caramuta S, Lee L, Ozata DM, Akcakaya P, Georgii-Hemming P, Xie H, Amini RM, Lawrie CH, Enblad G, Larsson C, et al. Role of microRNAs and microRNA machinery in the pathogenesis of diffuse large B-cell lymphoma. Blood Cancer J. 2013;3:e152. doi: 10.1038/bcj.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, Liu C, Kabar I, Schmidt HH, Zhang X, et al. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60:590–598. doi: 10.1016/j.jhep.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 32.Zhu DX, Zhu W, Fang C, Fan L, Zou ZJ, Wang YH, Liu P, Hong M, Miao KR, Liu P, et al. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33:1294–1301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 33.Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7:2152–2159. doi: 10.1158/1535-7163.MCT-08-0021. [DOI] [PubMed] [Google Scholar]
- 34.Bai H, Wei J, Deng C, Yang X, Wang C, Xu R. MicroRNA-21 regulates the sensitivity of diffuse large B-cell lymphoma cells to the CHOP chemotherapy regimen. Int J Hematol. 2013;97:223–231. doi: 10.1007/s12185-012-1256-x. [DOI] [PubMed] [Google Scholar]
- 35.Marques SC, Ranjbar B, Laursen MB, Falgreen S, Bilgrau AE, Bodker JS, Jorgensen LK, Primo MN, Schmitz A, Ettrup MS, et al. High miR-34a expression improves response to doxorubicin in diffuse large B-cell lymphoma. Exp Hematol. 2016;44(238–46):e2. doi: 10.1016/j.exphem.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 36.Alencar AJ, Malumbres R, Kozloski GA, Advani R, Talreja N, Chinichian S, Briones J, Natkunam Y, Sehn LH, Gascoyne RD, et al. MicroRNAs are independent predictors of outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin Cancer Res. 2011;17:4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Troppan K, Wenzl K, Pichler M, Pursche B, Schwarzenbacher D, Feichtinger J, Thallinger GG, Beham-Schmid C, Neumeister P, Deutsch A. miR-199a and miR-497 are associated with better overall survival due to increased chemosensitivity in diffuse large B-cell lymphoma patients. Int J Mol Sci. 2015;16:18077–18095. doi: 10.3390/ijms160818077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong HL, Zhou SW, Sun AH, He Y, Li J, Yuan X. MicroRNA197 reverses the drug resistance of fluorouracilinduced SGC7901 cells by targeting mitogenactivated protein kinase 1. Mol Med Rep. 2015;12:5019–5025. doi: 10.3892/mmr.2015.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Z, Zhou N, Han Q, Zhao L, Bai C, Chen Y, Zhou J, Zhao RC. MicroRNA-197 influences 5-fluorouracil resistance via thymidylate synthase in colorectal cancer. Clin Transl Oncol. 2015;17:876–883. doi: 10.1007/s12094-015-1318-7. [DOI] [PubMed] [Google Scholar]
- 40.Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, De Maria R. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ. 2014;21:774–782. doi: 10.1038/cdd.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Li F, Saha MN, Abdi J, Qiu L, Chang H. miR-137 and miR-197 induce apoptosis and suppress tumorigenicity by targeting MCL-1 in multiple myeloma. Clin Cancer Res. 2015;21:2399–2411. doi: 10.1158/1078-0432.CCR-14-1437. [DOI] [PubMed] [Google Scholar]
- 42.Swerdllow S, Campo E, Harris NL. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon: IARC Press; 2008. [Google Scholar]
- 43.Cheson BD, Pfistner B, Juweid ME, Gascoyne RD, Specht L, Horning SJ, Coiffier B, Fisher RI, Hagenbeek A, Zucca E, et al. Revised response criteria for malignant lymphoma. J Clin Oncol. 2007;25:579–586. doi: 10.1200/JCO.2006.09.2403. [DOI] [PubMed] [Google Scholar]
- 44.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, Schwartz M, Venables B. gplots: various R programming tools for plotting data. R package version 3.0.1; 2016. http://CRAN.R-project.org/package=gplots. Accessed 5 Dec 2017.
- 45.Mazumdar M, Glassman JR. Categorizing a prognostic variable: review of methods, code for easy implementation and applications to decision-making about cancer treatments. Stat Med. 2000;19:113–132. doi: 10.1002/(SICI)1097-0258(20000115)19:1<113::AID-SIM245>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 46.Paul J, Soujon M, Wengner AM, Zitzmann-Kolbe S, Sturz A, Haike K, Keng Magdalene KH, Tan SH, Lange M, Tan SY, et al. Simultaneous inhibition of PI3Kdelta and PI3Kalpha induces ABC-DLBCL regression by blocking BCR-dependent and -independent activation of NF-kappaB and AKT. Cancer Cell. 2017;31:64–78. doi: 10.1016/j.ccell.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, Lih CJ, Williams PM, Shaffer AL, Gerecitano J, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med. 2015;21:922–926. doi: 10.1038/nm.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wenzel SS, Grau M, Mavis C, Hailfinger S, Wolf A, Madle H, Deeb G, Dorken B, Thome M, Lenz P, et al. MCL1 is deregulated in subgroups of diffuse large B-cell lymphoma. Leukemia. 2013;27:1381–1390. doi: 10.1038/leu.2012.367. [DOI] [PubMed] [Google Scholar]
- 49.Sehn LH, Gascoyne RD. Diffuse large B-cell lymphoma: optimizing outcome in the context of clinical and biologic heterogeneity. Blood. 2015;125:22–32. doi: 10.1182/blood-2014-05-577189. [DOI] [PubMed] [Google Scholar]
- 50.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 51.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature. 2010;463:88–92. doi: 10.1038/nature08638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 53.Demchenko YN, Kuehl WM. A critical role for the NFkB pathway in multiple myeloma. Oncotarget. 2010;1:59–68. doi: 10.18632/oncotarget.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bard JD, Gelebart P, Anand M, Amin HM, Lai R. Aberrant expression of IL-22 receptor 1 and autocrine IL-22 stimulation contribute to tumorigenicity in ALK+ anaplastic large cell lymphoma. Leukemia. 2008;22:1595–1603. doi: 10.1038/leu.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lerman G, Sharon M, Leibowitz-Amit R, Sidi Y, Avni D. The crosstalk between IL-22 signaling and miR-197 in human keratinocytes. PLoS ONE. 2014;9:e107467. doi: 10.1371/journal.pone.0107467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang H, Su X, Yang M, Chen T, Hou J, Li N, Cao X. Reciprocal control of miR-197 and IL-6/STAT3 pathway reveals miR-197 as potential therapeutic target for hepatocellular carcinoma. Oncoimmunology. 2015;4:e1031440. doi: 10.1080/2162402X.2015.1031440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, Kohlhammer H, Xu W, Yang Y, Zhao H, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaque JP, Martinez N, Batlle-Lopez A, Perez C, Montes-Moreno S, Sanchez-Beato M, Piris MA. B-cell lymphoma mutations: improving diagnostics and enabling targeted therapies. Haematologica. 2014;99:222–231. doi: 10.3324/haematol.2013.096248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunleavy K, Pittaluga S, Czuczman MS, Dave SS, Wright G, Grant N, Shovlin M, Jaffe ES, Janik JE, Staudt LM, et al. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood. 2009;113:6069–6076. doi: 10.1182/blood-2009-01-199679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 61.Luminari S, Montanini A, Federico M. Anthracyclines: a cornerstone in the management of non-Hodgkin’s lymphoma. Hematol Rep. 2011;3:e4. doi: 10.4081/hr.2011.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee KW, Kim DY, Yun T, Kim DW, Kim TY, Yoon SS, Heo DS, Bang YJ, Park S, Kim BK, et al. Doxorubicin-based chemotherapy for diffuse large B-cell lymphoma in elderly patients: comparison of treatment outcomes between young and elderly patients and the significance of doxorubicin dosage. Cancer. 2003;98:2651–2656. doi: 10.1002/cncr.11846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fiugre S1. Hierarchical clustering of diffuse large B cell lymphoma cases and microRNA expression profiling with complete agglomerative (a), single agglomerative (b), Ward method (c), and divisive method (d).
Data Availability Statement
The datasets during and/or analyzed during the current study available from the corresponding author on reasonable request.