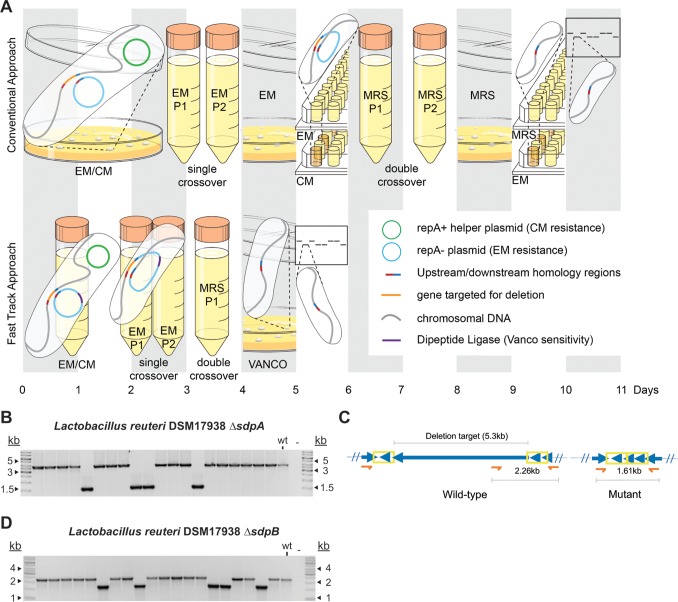
FIG 5.
Development of a fast-track genome editing approach. (A) Timeline comparison between the conventional approach and the fast-track liquid approach to generate DCO mutants in L. reuteri 17938. (Top panel) For the conventional approach, the suicide vector (with erythromycin resistance [Emr] and lacking repA [repA−]) containing homologous target regions was transformed into L. reuteri harboring a helper plasmid (with chloramphenicol resistance [Cmr] and repA [repA+]) on day 0. After electroporation and recovery, cells were plated on MRS medium containing erythromycin (EM) and chloramphenicol (CM) to select for cells containing both plasmids. At day 2, a single colony was transferred to MRS harboring erythromycin and cultured for a minimum of two passages followed by plating on MRS medium containing erythromycin. At day 5, colonies were tested for Emr and Cms in broth and were subjected to PCR to confirm SCO integration (not shown). To obtain DCO recombinants, cells were cultured in MRS medium for at least two passages and plated on nonselected plates. At day 9, extensive colony screening in broth was performed to identify Ems cells, i.e., cells in which a DCO event had occurred. Within the pool of Ems cells, PCR was performed to identify the desired genotype (day 11). (Bottom panel) For the fast-track approach, cells harboring the helper plasmid were transformed with the suicide vector encoding the dipeptide ligase (Ddl) enzyme. In bacteria that are intrinsically resistant to vancomycin (VANCO), expression of the dipeptide ligase yields cells that are more sensitive to vancomycin. After recovery, all cells were transferred to MRS broth containing erythromycin and chloramphenicol. After growth had been observed on day 2, cells were washed once in MRS medium and subcultured to MRS medium containing erythromycin for two passages. Cells then underwent one passage in MRS medium followed by plating on MRS medium containing vancomycin (days 3 and 4). The colonies that could grow in the presence of vancomycin had lost the suicide vector encoding Ddl and were subjected to PCR to identify recombinant genotypes. (Illustration by O'Reilly Science Art, LLC.) (B) Deletion of sdpA (2.2 kb) in L. reuteri DSM 17938. (C) Oligonucleotide design to identify in-frame deletion in sdpB locus. The oligonucleotide combination oVPL2072-oVPL2073-oVPL2694 can identify wild-type or deletion genotypes, yielding an amplicon of 2.2 or 1.6 kb, respectively. Yellow box, homologous region; orange arrow, oligonucleotides. (D) Deletion of sdpB (5.3 kb) in L. reuteri DSM 17938.