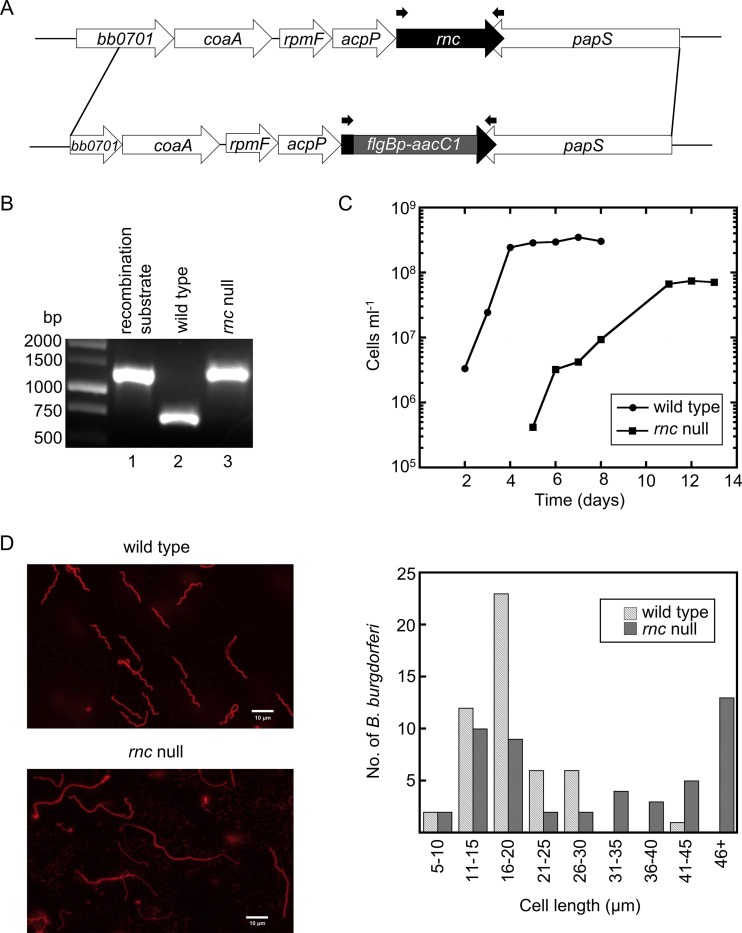
FIG 1.
A B. burgdorferi RNase III (rncBb) null mutant exhibits growth and morphological phenotypes. (A) Schematic of the genetic approach taken to construct the rncBb null mutant in B. burgdorferi. A gentamicin resistance cassette (flgBp-aacC1) replaced most of the chromosomal rncBb gene via allelic exchange by homologous recombination. Primers rnc 1F+NdeI and rnc 738R (Table 1), which were used to obtain PCR products shown in panel B, are indicated by the small black arrows above the rncBb gene and flgBp-aacC1 cassette. (B) Confirmation of an rncBb null mutant (in a B31 background). DNA from a B. burgdorferi transformant and controls were amplified by PCR, using primers flanking the insertion site, and the products were electrophoresed in a 1% agarose gel. Lane 1, cloned recombination substrate; lane 2, genomic DNA from wild-type B. burgdorferi; lane 3, genomic DNA from B. burgdorferi rnc null mutant. (C) B. burgdorferi wild type and the rncBb null mutant were inoculated in BSK II liquid medium at a cell density of 104 cell ml−1 and grown at 34°C until stationary phase. Cells were enumerated every 24 h, using a Petroff-Hausser counting chamber. (D) Microscopy images showing the cell morphology of the rncBb null mutant (lower panel) compared with that of wild-type B. burgdorferi (upper panel). Live B. burgdorferi cells were stained with a wheat germ agglutinin (WGA)-Alexa Fluor 594 conjugate and assayed by fluorescence microscopy. The length of 50 cells for each strain was measured as described by Lybecker et al. (54) and sorted into bins of 5-μm increments.