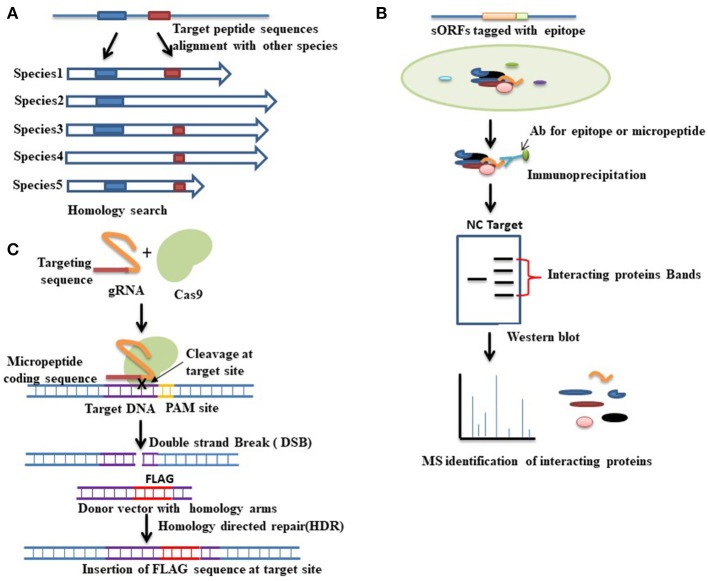
Figure 2.
Various approaches for functional characterization of micropeptides. (A) Evolutionary conservation of a peptide sequence is suggestive of functionality. Homology- based searching among species thus can be performed to identify whether the target peptide sequence shares any functional similarity with other proteins. Here the blue and red boxes indicate the conserved sequences among species. (B) Functional proteomics is a commonly used approach for identifying the interacting proteins of a target protein. In this method, first, immunoprecipitation is conducted by using an antibody (Ab) that is designed either against the epitope tagged with a target micropeptide or directly against the micropeptide. Western blot is then performed followed by mass spectrometry analysis to separate and identify the interacting proteins. Red brackets indicate the bands of interacting proteins that are separated by western blot analysis. A negative control (NC) denotes an empty vector that also runs for comparison. The nature of the interacting protein will thus provide clues about the function of the target micropeptide. (C) CRISPR-cas9 mediated gene editing approaches can also be used to check the coding potential of sORFs. To verify the coding potential, an epitope tag (FLAG) can be inserted at the downstream of the sORF into the endogenous locus. CRISPR-cas9 mediated gene editing is started by the recognition of the target site, which is mediated by a guide RNA (gRNA). Guide RNA guides the cas9 endonuclease to a specific location in the genome sequence, which is immediately adjacent to a protospacer adjacent motif (PAM). Upon recognition, the cas9 creates a double strand break (DSB) at the target site. This DSB can then be repaired either by non-homologous end joining (NHEJ) or by homology directed repair (HDR). HDR is used to insert an epitope tag at the target site where a donor vector with homology to the targeted locus must be provided. The donor vector must contain the epitope tag that has to be knocked-in at the target site. Expression of the engineered fusion protein can then be verified by western blot analysis.