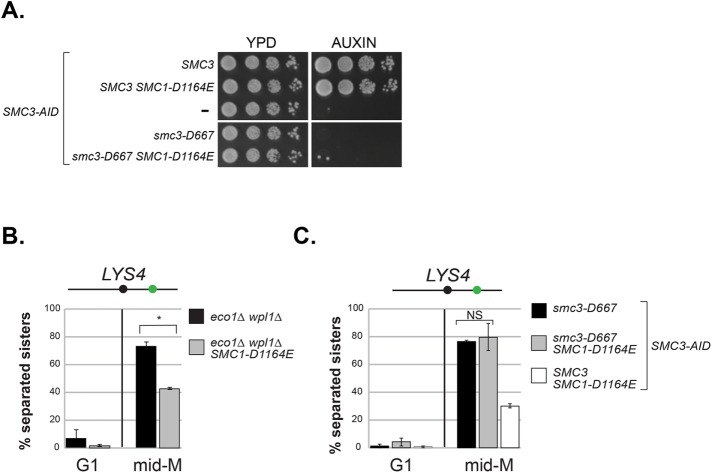
FIGURE 6:
The SMC1-D1164E mutation fails to suppress the inviability or cohesion defect of smc3-D667. (A) smc1-D1164E failed to restore viability to smc3-D667 cells. Haploid strains SMC3 SMC3-AID (BRY474), SMC3 SMC3-AID SMC1-D1164E (BRY832), SMC3-AID (VG3651-3D), smc3-D667 SMC3-AID (BRY482), and smc3-D667 SMC3-AID SMC1-D1164E (BRY833) were grown to saturation in YPD and then plated as 10-fold serial dilutions onto YPD alone (YPD) or containing 0.75 mM auxin (YPD + auxin) and incubated 2 d at 23°C. (B) SMC1-D1164E suppresses cohesion loss of the eco1∆ wpl1∆ mutant in mid–M phase–arrested cells. Haploid strains eco1∆ wpl1∆ (VG3503-4A) and SMC1-D1164E eco1∆ wpl1∆ (VG3575-2C) grown as described in Figure 2A. Cells from G1 and mid–M phase arrest were fixed and processed and scored for cohesion loss at the CEN-distal LYS4 locus. *p < 0.05 (t test, two-tailed). (C) SMC1-D1164E fails to suppress cohesion loss of smc3-D667 cells. Haploid strains smc3-D667 SMC3-AID (BRY482), smc3-D667 SMC3-AID SMC1-D1164E (BRY833), and SMC3 SMC3-AID SMC1-D1164E (BRY832) cells were grown according the regimen in Figure 2A and processed to assess cohesion loss at the CEN-distal LYS4 locus as described in B. For both B and C, the percentage of cells with two GFP foci (sister separation) were derived from two independent experiments. An amount of 100–200 cells was scored per sample at each time point. Error bars represent SD. NS = not significant (p = 0.74; t test, two-tailed).