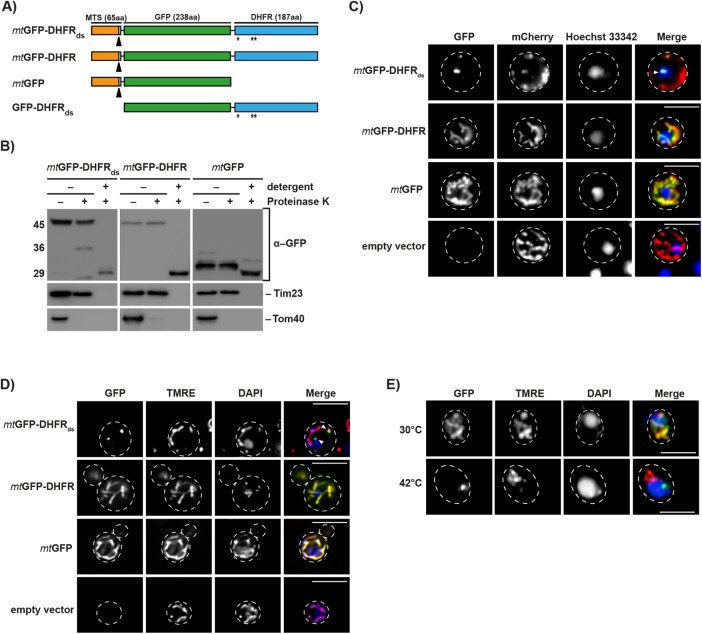
FIGURE 1:
Expression and localization of aggregation-prone mitochondrial reporter proteins. (A) Schematic depiction of the destabilized reporter construct mtGFP-DHFRds and the corresponding control constructs mtGFP-DHFR, mtGFP, and GFP-DHFRds. Indicated are the MTS (orange), the GFP domain of A. victoria (green), and the DHFR domain of Mus musculus. Destabilizing mutations in the DHFR domain are indicated (*). Cleavage site for mitochondrial processing peptidase is indicated by a black arrow. (B) Treatment of intact and lysed mitochondria containing destabilized reporter or control proteins with 2.5 µM proteinase K. Mitochondria were lysed by treatment with 0.5% Triton X-100. Proteinase K digestion was performed for 20 min at 4°C. Reporter proteins and control proteins of the outer (Tom40) and inner mitochondrial membrane (Tim23) were detected by Western blot. (C) Analysis of aggregate-containing live cells by fluorescence microscopy. Mitochondria-targeted destabilized GFP fusion proteins or controls (green) were expressed in yeast cells for 5 h. Cells also constitutively expressed the fusion protein b2(167)∆-mCherry (red) as a mitochondrial marker protein. Nuclei were visualized by staining with Hoechst-33342 (blue). IMiQ structures are indicated with a white arrow. Scale bar: 10 µm. (D) Cells expressing destabilized GFP-reporter or control proteins for 5 h were analyzed as above. Mitochondrial inner membrane potential (Δψ) was visualized by staining with TMRE (red) and nuclei stained with DAPI (blue). Scale bar: 10 µm. (E) Immunofluorescence of cells after expression of the folded control protein mtGFP-DHFR (green) for 5 h and subsequent incubation at the indicated temperatures for 4 h. Mitochondria were stained with TMRE and nuclei were visualized by staining with DAPI (blue). Scale bar: 10 µm.