Figure 1.
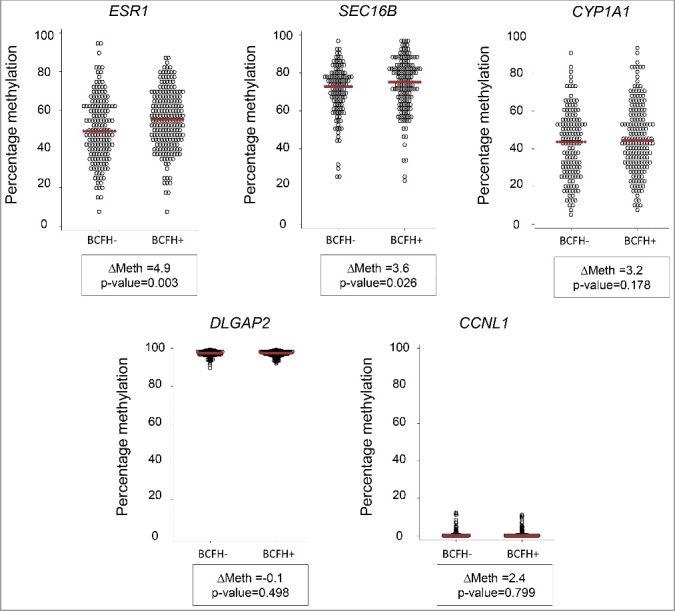
Methylation differences between BCFH+ and BCFH- girls in ESR1, SEC16B, CYP1A1, DLGAP2, and CCNL1. Net CpG methylation values (percent) in BCFH- girls and BCFH+ girls in 5 gene regions examined in WBC DNA. The red line represents the median methylation. ESR1, SEC16B, and CYP1A1 showed high inter-individual variability of the methylation level, while the distribution of methylation levels of DLGAP2 and CCNL1 did not. For the CYP1A1, DLGAP2 and CCNL1, there is no difference between BCFH+ and BCFH- girls, while for ESR1 and SEC16B, there are small but statistically significant differences. Importantly, the range of methylation values for these loci is wide, suggesting the presence of hap-ASM, in which one local haplotype acts in cis and dictates a low methylation level while another haplotype dictates high methylation.
