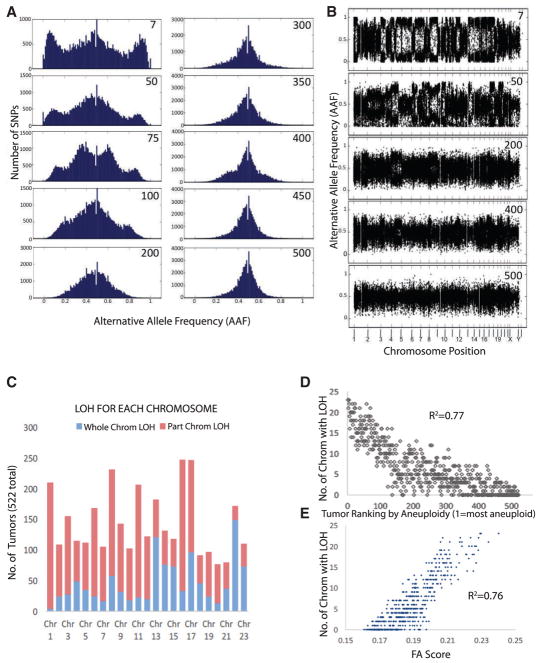
Figure 1. Measuring the Aneuploidy in Tumors by the Broadening of Allelic Frequency Ratios Calculated from Sequencing Data.
(A) 10 examples of FA histograms, each portraying the allelic frequencies of the heterozygous SNPs of human breast tumors. The number in the top right corner of each plot represents the FA ranking based on the broadening of the allele frequency peaks, with 1 being the widest peak and 510 the narrowest peak. Note that the lower the number, the broader the central peak of allelic frequencies.
(B) Replots of 5 of the tumors from (A) showing the allele frequency ratios along chromosome positions corresponding to coordinates in the GRCh37 genome (so each chromosome runs p arm to q arm). Note that the LOH events that separate the central peaks into two often span large segments of chromosomes or whole chromosomes, which is consistent with chromosome missegregation driving the LOH event.
(C) Plot of the frequency of whole (blue) or partial (red) chromosome LOH for each chromosome.
(D and E) The number of chromosomes with an LOH event in breast tumors correlates with the ranking (D) and FA score (E) of the tumors. R2 values were calculated by fitting to a second-order polynomial curve in Excel. Our method of LOH quantification is summarized in Figure S5.