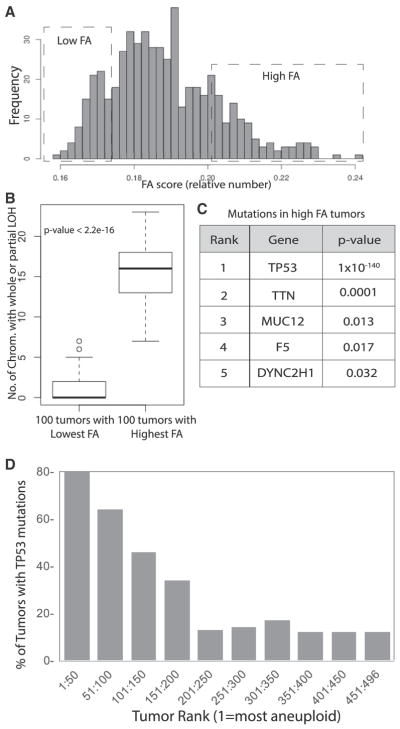
Figure 2. Identification of Genes Mutated in Aneuploid Tumors.
(A) Histogram showing the distribution of FA scores of the 522 analyzed human breast tumors. The boxes highlight the distributions of the 100 highest and lowest scoring tumors.
(B) The number of chromosomes with LOH events was compared in the 100 highest and lowest aneuploid tumors to demonstrate that the analysis stratifies tumors by aneuploid status. The p value was generated by Welch two-sample t test in R.
(C) All genes significantly mutated in the high aneuploid tumor sets were identified by comparing the sequence data from the 100 highest and lowest ranked tumors using the VAAST program. The p values were calculated by VAAST.
(D) p53 mutations are correlated with functional aneuploidy in the 250 top ranked tumors.