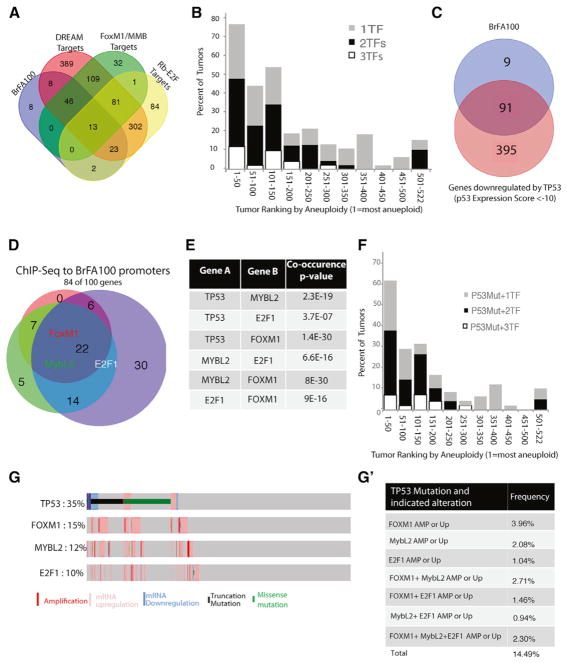
Figure 4. Mutations in TP53 and Overexpression of E2F1, MYB2L, and FOXM1 Are Highly Associated in Breast Tumors.
(A) The overlap of BrFA100 and target genes of mitotic transcriptional regulation complexes DREAM, FoxM1-MuvB/MMB, and Rb-E2F.
(B) The percentage of tumors in each group of 50 (ranked by aneuploid status) with 1, 2, or 3 of the transcription factors MYBL2, FOXM1, and E2F1.
(C) Venn diagram of the overlap of the BrFA100 with the top 400 genes downregulated by TP53 (p53 expression score of less than −10, as listed in Fischer et al., 2016).
(D) Venn diagram to show the overlap of ChIP-seq datasets for E2F1, MYB2L, and FOXM1 with the BrFA100 list. Gene lists are shown in Table S3.
(E) Association p values of TP53, E2F1, MYB2L, and FOXM1 as individual pairs. p values were obtained through Fisher exact tests with Benjamini-Hochberg multiple test corrections.
(F) The percentage of tumors in each group of 50 that have a TP53 mutation and 1, 2, or 3 overexpressed transcription factors.
(G) Association of TP53, MYBL2, E2F1, and FOXM1 in 960 human breast tumors of the TCGA. Plots were generated at the cBioPortal (www.cbioportal.org). (G’) The percentage of the 960 tumors with a TP53 mutation and either an amplification (AMP) of the gene as defined by a positive GISTIC score or an upregulation (Up) of the mRNA as defined by a Z score > 2. TF, transcription factor.
See also Table S3.