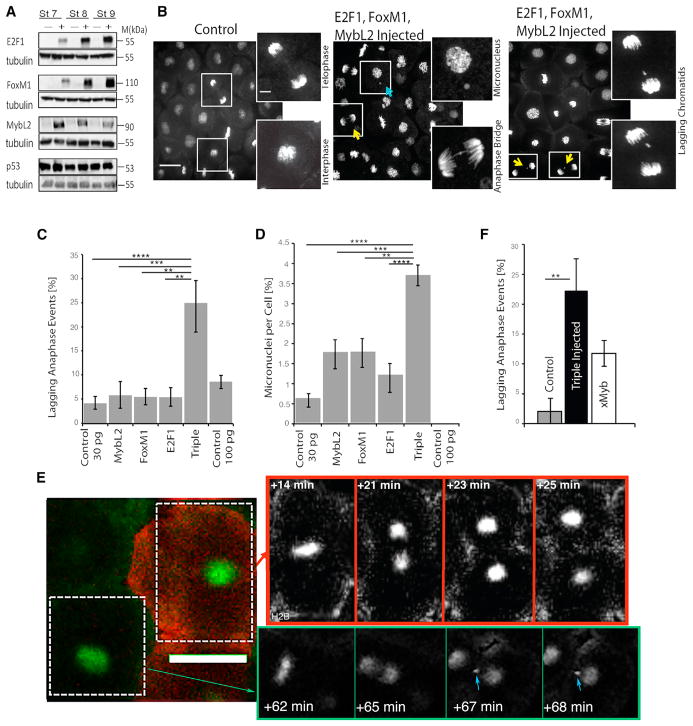
Figure 5. Overexpression of hE2F1, hFOXM1, and hMYBL2 Is Sufficient to Generate CIN Phenotypes in Xenopus Embryos.
(A) 2-cell-stage embryos were injected with either RNA containing stop codon after 33 nt (−) or functional hE2F1, hFoxM1, and hMybL2 (+), as detected by western blot.
(B) Representative images of TOPRO-stained normally dividing animal caps and two of the most common CIN phenotypes seen in triple-overexpressing embryos. Yellow arrows indicate a lagging chromatid; blue arrow indicates a micronucleus.
(C and D) Quantification of lagging chromatids (C) and micronuclei (D) in control, triply overexpressing, or singly injected embryos through fixed-animal cap analysis.
(E) Representative time-lapse series of an animal cap expressing H2B:GFP with normally dividing control cells (co-injected with Ruby-Dextran) or CIN-like phenotypes seen in neighboring triple-overexpressing cells. Blue arrows indicate abnormal divisions seen as lagging chromatids and micronuclei. Time points chosen to show anaphase events.
(F) Quantification of lagging chromatid events as seen in time-lapse videos of control embryos, triply overexpressing embryos, and overexpression of only xMYBL2. Full supplemental videos are available upon request. Scale bars represent 40 μm in all images. **p < 0.01; ***p < 0.001; ****p < 0.0001, one-way ANOVA and Bonferroni post-test statistics,. Error bars represent ±SEM. 8 hpf, 8 hr post-fertilization.