Figure 5.
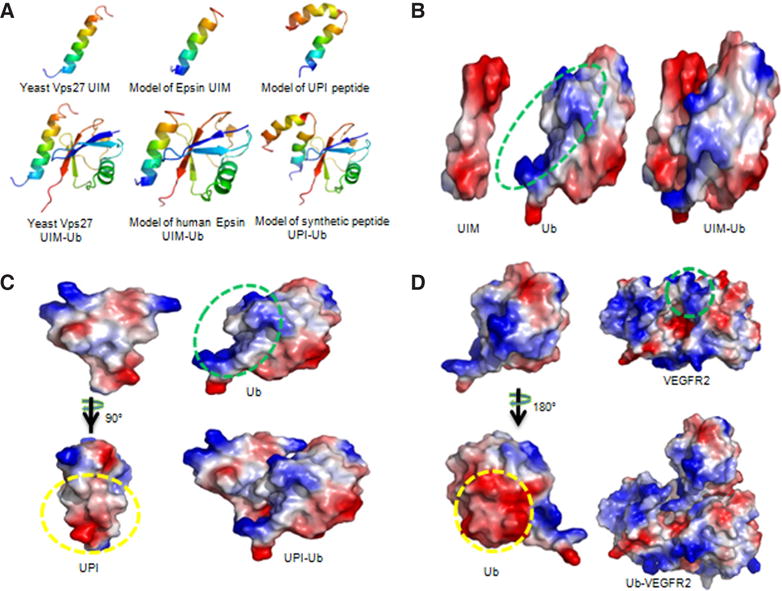
Models of UIM-Ub, UPI-Ub, and Ub-VEGFR2 complexes. (A) Ribbon diagrams show yeast Vps27 UIM interaction with Ub (left), and models of human epsin UIM-Ub complex (middle) and UPI-Ub complex (right). The NMR structure of yeast Vps27 and X-ray structure of Ub were taken from the Protein Data Bank, entries 1Q0W and 3JVZ, respectively. The top scoring models of epsin UIM and UPI (top panel) were selected and docked into Ub. The models with high scores and good topologies are shown in the bottom panel. Yeast Vps27 UIM, epsin UIM, and UIM of UPI peptide interact with Ub in a highly similar manner (bottom panel). Structures are multi-colored, with the N and C termini denoted in blue and red, respectively; (B-D) Electrostatic surface representations of UIM-Ub, UPI-Ub, and Ub-VEGFR2 complex models. Red and blue represent negative and positive potential, respectively. The proposed binding surfaces with negatively charged amino acids are indicated by yellow circles. The green circles highlight the proposed interaction surfaces of positively charged amino acids. The figures were prepared using PyMol (Schrödinger, Inc, Cambridge, MA). Note: Figure 5A is adapted with permission from ref. 19. Copyright 2015 ASCI
