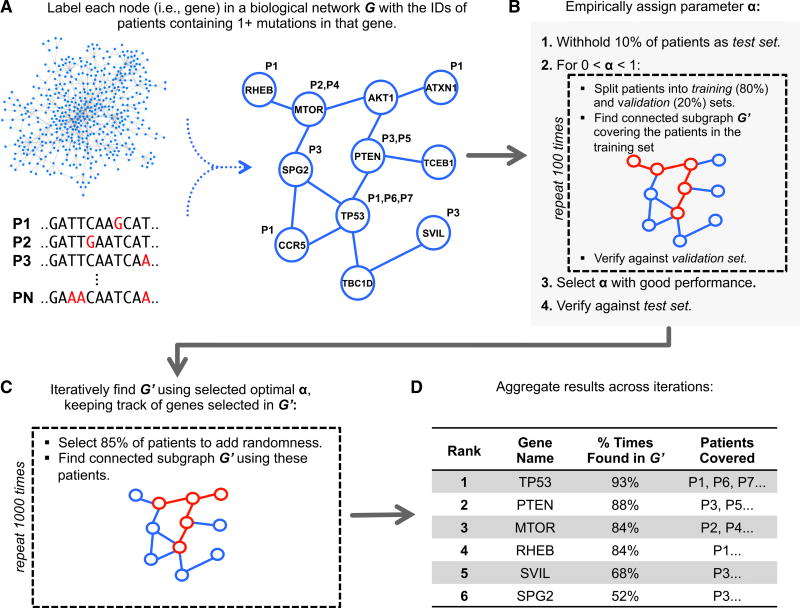
Figure 1. Overview of Our Approach.
(A) Somatic mutations are mapped onto a protein-protein interaction network. Each node is associated with the set of individuals whose cancers have mutations in the corresponding gene. The overall goal is to select a small connected subnetwork such that most individuals in the cohort have mutations in at least one of the corresponding genes (i.e., are “covered”).
(B) nCOP automatically selects a value for the parameter α by performing a series of cross-validation tests. First, 10% of the individuals are withheld as a test set. Next, the remaining individuals are repeatedly and randomly split into two groups, a training set (80%) and a validation set (20%). For each split, the nCOP search heuristic is run for 0 < α < 1 using the individuals comprising the training set. An α is selected to obtain high coverage of the individuals in the validation sets while maintaining similar coverage on the training sets (i.e., not overfitting to the training sets). Coverage of individuals in the initially withheld test set is also calculated and confirmed to be similar to the validation sets.
(C) Once α is selected, to avoid overfitting on the entire dataset, nCOP is run 1,000 times using random subsets of 85% of the individuals.
(D) Finally, the subnetworks output across the runs are aggregated and candidate genes are ranked by the number of the times they appear across these subnetworks.