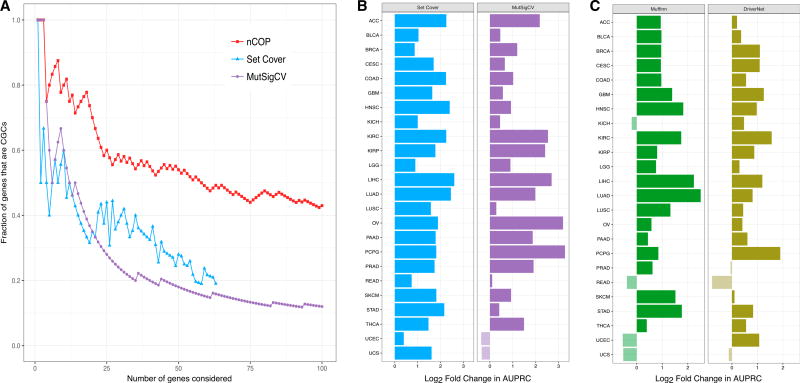
Figure 3. nCOP Is More Successful than Other Methods in Identifying Known Cancer Genes.
(A) Our network-based algorithm nCOP, a set cover version of our algorithm that ignores network information, and MutSigCV, a frequency-based approach, are compared on the KIRC dataset. nCOP ranks genes based on how frequently they are output, and MutSigCV ranks genes by q values. The set cover approach is run for increasing values of k until all patients are covered. For each method, as an increasing number of genes are considered, we compute the fraction that correspond to CGCs. Over a range of thresholds, our algorithm nCOP outputs a larger fraction of CGC genes than the other two approaches.
(B) Comparison of nCOP to two network-agnostic methods across 24 cancer types. For each cancer type, we compute AUPRCs for nCOP, the set cover approach, and MutSigCV, using their top 100 predictions. We give the log2 ratios of nCOP’s AUPRCs to the other methods’ AUPRCs. Our approach nCOP outperforms the set cover approach on all 24 cancers, and MutSigCV on 22 of the 24 cancer types.
(C) Comparison of nCOP with two network-based methods, Muffinn and DriverNet, across 24 cancer types. Our approach nCOP outperforms Muffinn and DriverNet on 20 and 21, respectively, of the 24 cancer types.