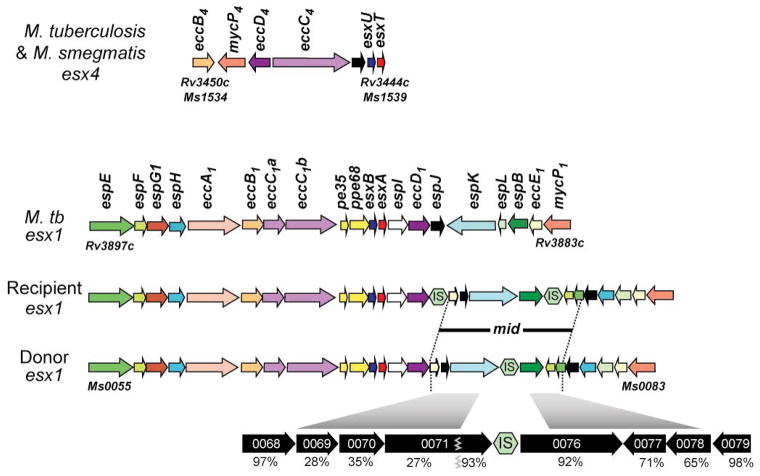
Fig. 4.
esx4 loci share common genes necessary for ESX function. The seven-gene core locus of esx4 is conserved in gene content and order in most mycobacteria, including M. tuberculosis and M. smegmatis donor and recipient strains (top panel). The esx1 locus is more complex and is more heterogeneous in the composition of genes near its 3′ boundary, particularly where the mating identity (mid) genes are located in M. smegmatis (bottom panel). The mid genes are expanded at the bottom to show the level of amino acid identity between donor and recipient strains (IS represents insertion sequence elements). The genes are colour coded to indicate orthologous genes in each ESX system. The M. smegmatis (Ms) and M. tuberculosis (Rv) gene numbers are given to indicate the location of each locus in its respective genome. The gene names follow the classification according to (Bitter et al., 2009). ecc genes are conserved components of all esx loci and are thought to encode core machinery. esp genes are specific to a locus, in this case esx1, and are thought to mediate locus-specific functions. EspB, PE35 and PPE68 are known to be secreted by ESX-1 (and figure 5). EsxB and EsxA form the heterodimer secreted by the ESX-1 system. The equivalent proteins in esx4, EsxU and EsxT, have yet to be shown to be secreted.