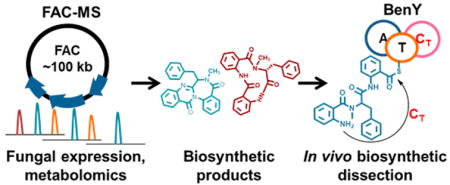
Abstract
The benzodiazepine benzomalvin A/D is a fungally derived specialized metabolite and inhibitor of the substance P receptor NK1, biosynthesized by a three-gene nonribosomal peptide synthetase cluster. Here, we utilize fungal artificial chromosomes with metabolomic scoring (FAC-MS) to perform molecular genetic pathway dissection and targeted metabolomics analysis to assign the in vivo role of each domain in the benzomalvin biosynthetic pathway. The use of FAC-MS identified the terminal cyclizing condensation domain as BenY-CT and the internal C-domains as BenZ-C1 and BenZ-C2. Unexpectedly, we also uncovered evidence suggesting BenY-CT or a yet to be identified protein mediates benzodiazepine formation, representing the first reported benzodiazepine synthase enzymatic activity. This work informs understanding of what defines a fungal CT domain and shows how the FAC-MS platform can be used as a tool for in vivo analyses of specialized metabolite biosynthesis and for the discovery and dissection of new enzyme activities.
Graphical Abstract
Estimates of fungal biodiversity on Earth range from 1 to 5 million species, of which approximately 100000 have been cataloged in various strain collections.1 More than half of these uncatalogued fungi are predicted to be ascomycetes, whose genomes encode tremendous biosynthetic potential, with typical examples possessing between 50 and 100 distinct biosynthetic gene clusters encoding the enzyme machinery to produce unique specialized metabolites (alternately called “secondary metabolites” or “natural products”).1–4 If one conservatively assumes that half of all fungi are ascomycetes and each has 50 biosynthetic gene clusters, then there are between 25 and 125 million fungal specialized metabolite biosynthetic gene clusters on Earth, of which only a very small minority (≪1%) has been studied. In addition to the likelihood that these clusters encode numerous valuable new small molecules in the tradition of such drugs as penicillin, lovastatin, and cyclosporine,5,6 this genetic repository has the potential to reveal new functional insights into biosynthesis, metabolic pathway engineering, and enzymology.
Despite this large biochemical potential, the study of specialized metabolites from fungi has lagged behind bacteria and plants, in part because of difficulties either culturing or genetically manipulating many fungal species in the lab. Recently, we reported fungal artificial chromosomes with metabolomic scoring (FAC-MS) as a robust new platform for the discovery of fungal specialized metabolites and their biosynthetic gene clusters.7 FAC-MS enables specialized metabolite discovery through high-throughput capture of intact fungal biosynthetic gene clusters from genomic DNA in an Escherichia coli–Aspergillus nidulans shuttle vector called a FAC,8 followed by gene cluster heterologous expression in A. nidulans and detection and analysis of small molecule products by untargeted metabolomics and cheminformatics using high-resolution mass spectrometry. Because the FAC serves as a shuttle vector, FAC genetic recombineering in E. coli can be subsequently used for facile “gene cluster editing” of biosynthetic gene clusters, followed by transformation into A. nidulans to enable rapid targeted metabolomic and genetic validation of gene cluster–metabolite relationships.
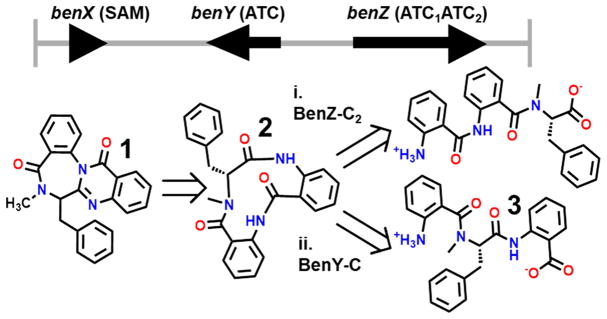
Using this approach, we reported for the first time the biosynthetic gene cluster for the benzodiazepine specialized metabolites, benzomalvin A and D, an atropisomeric pair of metabolites in conformational equilibrium [herein collectively called benzomalvin A/D (1) (see Scheme 1)].7 Benzomalvin A/D was first discovered from a Penicillium species and shown to antagonize the human NK1 receptor, inhibiting the effects of substance P;9 benzomalvins also inhibit the human enzyme 2,3-indoleamine dioxygenase, a potential therapeutic target in pathologies ranging from autoimmune disorders to Alzheimer’s disease.10 The benzomalvin biosynthetic gene cluster was discovered using FAC-MS in Aspergillus terreus (ATCC 20542) and consists of three genes: one putative SAM-binding methyltransferase benX and two nonribosomal peptide synthetase (NRPS) genes benY and benZ (Figure 1). On the basis of bioinformatic A-domain analysis,11,12 the A-domain of BenY and the first A-domain of BenZ are both predicted to incorporate the anthranilate monomer (Anth), which is a known fungal biosynthetic building block of benzodiazepines and quinazolinones.13 The second A-domain of BenZ is predicted to incorporate a large hydrophobic amino acid such as Phe, but not Anth. FAC deletions of benZ and benY revealed both genes to be necessary to benzomalvin biosynthesis.7
Scheme 1. Benzomalvin Products and Intermediatesa.
aThe benzodiazepine core is colored orange, while the N-methyl functionality is colored blue and the desmethyl amide red. Legend: 1, benzomalvin A/D; 2, 11-member macrocyclic tripeptide precursor; 3, linear anthranilate-N-methyl-phenylalanine-anthranilate (Anth-NmPhe-Anth) tripeptide released by spontaneous hydrolysis of the phosphopantetheinylated precursor; 4, desmethyl benzomalvin A/D.
Figure 1.
Two possible benzomalvin retrobiosyntheses. The benzomalvin biosynthetic machinery consists of NRPS enzymes BenY and BenZ, as well as methyltransferase BenX. Retrobiosynthetic analysis of benzomalvin combined with the biosynthetic rules of colinearity predicts two possible linear tripeptide precursors that would arise from different C-domain activities: Anth-Anth-NmPhe that would require CT activity by BenZ-C2 and Anth-NmPhe-Anth that would require CT activity by BenY-C.
On the basis of the precedent set by biosynthetic studies of the chemically related specialized metabolites asperlicin C and D,13 retrobiosynthetic analysis of benzomalvin A suggests biosynthesis proceeds through formation of an NRPS-tethered linear tripeptide intermediate composed of two Anth residues and one N-methylphenylalanine (NmPhe) residue, which is then cyclized to form an 11-member macrocycle intermediate (2). Based on the asperlicins, 2 is then predicted to readily undergo a non-enzymatic transannulation reaction to form the benzodiazepine product, benzomalvin A/D (Figure 1). In the case of the asperlicins, two regioisomers with similar total abundances, asperlicins C and D, are formed from the final non-enzymatic transannulation step. In the case of the benzomalvins, however, regioisomerism is not observed. This indicates that within benzomalvin biosynthesis transannulation is regioselective, raising the possibility of a specific enzyme that catalyzes benzodiazepine formation.
Though the asperlicins and benzomalvins are chemically related, their gene clusters differ significantly in that asperlicins C and D are formed through the action of a single, iterative NRPS enzyme with two A-domains and two condensation domains (C-domains), whereas the benzomalvins are produced by a cluster encoding two separate NRPS enzymes with a total of three A-domains and three C-domains (Figure 1). Thus, A-domain analysis and colinearity14 suggest that benzomalvin A/D biosynthesis proceeds through a linear tripeptide with an Anth-Anth-NmPhe or Anth-NmPhe-Anth sequence. On the basis of this information, the specific chemical activities of BenY and BenZ C-domains cannot be inferred, nor can the order in which BenY and BenZ act. Of the three benzomalvin C-domains, two are expected to be internal, canonical C-domains, each catalyzing a peptide bond formation, while the third is expected to be a “terminal C-domain” (CT) that catalyzes the final cyclization and release to form 2.15,16 On the basis of phylogenetic analysis, BenZ-C1 clearly groups with internal C-domains. However, both BenY-C and BenZ-C2 cluster with CT domains and therefore cannot be differentiated by bioinformatics (Figure S1).
Herein, we applied a molecular genetic approach and introduced the concept of FAC domain deletants, to delete the C-domains of benY and benZ from a FAC containing the full benzomalvin gene cluster. The ΔbenZ-C2 and ΔbenY-C FACs were then analyzed by heterologous expression in A. nidulans and subsequent targeted metabolomics to directly determine the effect of these deletions on benzomalvin biosynthesis in fungi and to determine the role of each C-domain in benzomalvin biosynthesis. We report that BenY-C acts as the terminal C-domain, indicating the precise sequence of events of benzomalvin peptide biosynthesis and also supporting future bioinformatics efforts to predict and identify fungal CT domains. We also uncover evidence of a benzodiazepine synthase enzyme, which may aid future efforts to discover new benzodiazepines by genome mining, or to produce them by semisynthetic or bioengineering methods. More broadly, in addition to informing what defines a fungal CT domain, this work demonstrates how the FAC-MS pipeline with targeted domain-level deletions can be efficiently used to dissect metabolite biosynthesis, and for the discovery of novel enzymatic activities.
MATERIALS AND METHODS
FAC Clone AtFAC9J20 and FAC DNA Preparation
The shuttle BAC/FAC clone of AtFAC9J20 was maintained in the E. coli strain DH10B replicator and transformed into Red/ET-inducible E. coli strain SW012. Each FAC including the deletion mutants had high-quality DNA prepared from 200 mL each of FAC LB cultures with a FAC/BAC DNA preparation kit (Intact Genomics), and each FAC DNA pellet was dissolved in 200 μL of 10 mM Tris-HCl (pH 8.0) for subsequent A. nidulans transformation and specialized metabolite screening.
Gene Cluster Editing via FAC and Production of FAC Deletants
Red/ET tools have been developed for efficient large DNA- or BAC-based recombineering and transgenic animal models.20–22 The FAC recombineering protocol is a one-step targeting system using either GalK or antibiotic selection. Domain deletions were made in the same manner as previously reported for whole gene deletions.7 Briefly, to delete the benY-C or benZ-C2 domain, a pair of polymerase chain reaction (PCR) primers of the kanamycin resistance gene (21 bases forward and 18 bases reverse shown in Table S2 as lowercase letters) with an additional 50 bp homology sequence (uppercase letters shown in Table S2) flanking the appropriate domain of either benY-C or benZ-C2 were used to generate the GalK-PCR or Kan-PCR product, respectively, each of which was gel-purified. FAC DNA was transformed into Red/ET-inducible E. coli strain SW012. Induced SW102/FAC electro-competent cells (125 μL for five reactions) were made from a 50 mL culture, and the purified GalK- or Kan-PCR product (~30 ng) was used to transform the induced SW102/FAC competent cells (25 μL) before plating. The recombineered FAC colonies were then selected on either minimal medium with galactose or LB medium with kanamycin and confirmed by PCR, restriction digestion, pulse field gel electrophoresis, and sequencing (if needed). The deletions were completed with the primers listed in Table S2.
FAC Transformation, Growth, and Extraction
All transformations of FACs into A. nidulans and subsequent growth and extractions from transformants were performed as previously reported.7
Condensation Domain Bioinformatic Analysis
Minimum Information about a Biosynthetic Gene Cluster (MIBiG) sequences were downloaded and filtered for ascomycete NRPS gene clusters.23 Excised condensation domain sequences (matching Pfam 00668; n = 286) were aligned using MUSCLE.24 A maximum-likelihood tree was constructed from aligned sequences in MEGA 7 using a bootstrap method phylogeny test.25 Internal and terminal condensation domain sequences were separately realigned using MUSCLE, and profile hidden Markov models were created using the HMMER web server.26
Metabolomics Analysis
Targeted metabolomic analysis was performed using previously described methods.7 Briefly, dried metabolite extracts were resuspended in 50% acetonitrile to a concentration of 2 mg/mL and analyzed by injecting 25 μL on a Phenomenex Luna C-18(2) reverse phase column with dimensions of 2 mm × 150 mm, 3 μm dp, using an Agilent 1200 series high-performance liquid chromatograph. The gradient was as follows: 2% B at 0 min, 70% B at 35 min, and 98% B at 54 min, with a flow rate of 200 μL/min (buffer A being H2O with 0.1% formic acid and buffer B being acetonitrile with 0.1% formic acid). The column eluent flowed into an Ion Max electrospray ionization source connected to a Q-Exactive mass spectrometer in positive mode, with top 5 data-dependent collection with an intact mass scan range of 150–2000 Th and a tandem mass spectrum isolation window of ±1 Th. The exclusion window was set at 60 s. The resolution was 70000 at m/z 200, and the higher-energy collision cell was set to a NCE value of 30. Metabolite peak areas were integrated, normalized relative to the total ion current, and averaged across biological quadruplicate metabolite extracts to determine metabolite abundances.7 Error bars for metabolite abundances were derived from the standard deviation.
RESULTS AND DISCUSSION
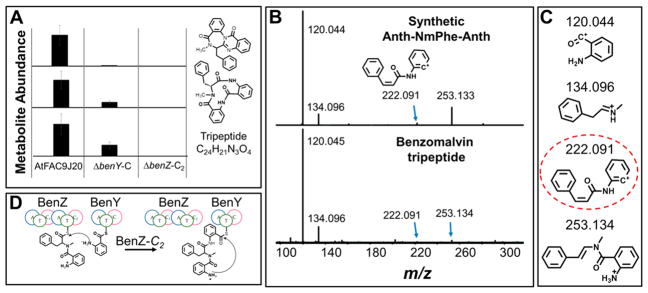
To determine the roles of individual NRPS domains in benzomalvin biosynthesis, FAC genetic recombineering was used to produce deletions of the benY-C and benZ-C2 domains, leading to constructs AtFAC9J20-ΔbenY-C and AtFAC9J20-ΔbenZ-C2, respectively. These constructs were transformed into A. nidulans and their secreted metabolomes analyzed by untargeted liquid chromatography and mass spectrometry (LC–MS) to determine the abundances of benzomalvins and their precursors. In the case of AtFAC9J20 without any deletions, 1, 2, 4, and an unidentified compound with a molecular formula matching that of 3 were readily detected. However, the relative abundances of each of these were significantly decreased in the case of the AtFAC9J20-ΔbenY-C metabolome. These metabolites could not be detected at all from the AtFAC9J20-ΔbenZ-C2 metabolome (Figure 2A).
Figure 2.
BenY-C is the CT domain in benzomalvin biosynthesis. (A) The relative abundances of benzomalvin A/D (1), the macrocyclic precursor (2), and the linear tripeptide precursor were determined by LC–MS in AtFAC9J20, AtFAC9J20-ΔbenY-C, and AtFAC9J20-ΔbenZ-C2. The product from AtFAC9J20-ΔbenY-C suggests BenY-C as the CT domain because deletion of an internal C-domain would likely fully block biosynthesis (also see Figure S2). (B) MS2 fragmentation spectrum for the free tripeptide intermediate of benzomalvin biosynthesis compared to that of a synthetic standard of 3, showing the two parent ions share identical fragments, including the diagnostic ion at m/z 222.091 that would not be produced by an Anth-Anth-NmPhe tripeptide. (C) MS2 fragment ions observed in both the synthetic Anth-NmPhe-Anth standard and the benzomalvin linear tripeptide precursor. The dashed circle highlights the diagnostic fragment for the Anth-NmPhe-Anth peptide. (D) Benzomalvin biosynthesis proceeds through a linear Anth-NmPhe-Anth tripeptide intermediate, with BenZ-C2 catalyzing the second peptide condensation.
On the basis of biosynthetic colinearity, there are two credible biosynthetic pathways by which 1 and 2 could be produced (Figure S2). In pathway i, BenY-C acts as the first internal C-domain, while in pathway ii, BenY-C acts as the CT domain. Because internal C-domains are required to facilitate protein–protein interactions to allow condensation reactions between separate NRPS enzymes, deletion of an internal C-domain is likely to fully block biosynthesis. On the other hand, deletion of the CT domain is much more likely to be bypassed through non-enzymatic attack of the locally tethered nucleophile on the phosphopantetheinylated intermediate’s thioester bond. Thus, production of 1 and 2 by AtFAC9J20-ΔbenY-C, albeit at a level lower than that of AtFAC9J20, suggests that benY-C encodes the CT domain and that benzomalvin is biosynthesized through pathway ii. In further support of this, 1 and 2 were not detected from AtFAC9J20-ΔbenZ-C2, indicating BenZ-C2 is not readily bypassed, consistent with it having a role as an internal C-domain.
Pathways i and ii could also be differentiated by the identification of unique intermediates. Benzomalvin biosynthesis by pathway ii would lead to production of a phosphopantetheinylated tripeptide intermediate with an Anth-NmPhe-Anth sequence, while pathway i would produce Anth-Anth-NmPhe. Previously, we reported detection of a benzomalvin-related metabolite with a molecular formula that corresponds to the free hydrolysis product of either an Anth-NmPhe-Anth (3) or an Anth-Anth-NmPhe intermediate, suggesting that some of the phosphopantetheinylated tripeptide precursor is non-enzymatically hydrolyzed and released from the NRPS assembly line. However, the tripeptide could not be sequenced by tandem mass spectrometry because of its low abundance.
To test if this tripeptide matches the sequence of the Anth-NmPhe-Anth tripeptide (3) predicted by pathway ii, we prepared a synthetic standard of 3 and compared its tandem mass spectrum to that recorded for the free tripeptide product (Figure 2B,C). The four most abundant fragments obtained from 3 were also detected with high mass accuracy from the free tripeptide product and are shown in Figure 3C. Of these fragments, the species at m/z 222.091 is an expected product from fragmentation of 3, but not from an Anth-Anth-NmPhe tripeptide. Thus, tandem mass spectrometry analysis of the free tripeptide and synthetic 3 suggests that the sequence of the tripeptide is Anth-NmPhe-Anth, which is consistent with biosynthetic pathway ii, but not pathway i (Figure S2). Together, these data indicate that BenZ-C2 acts as the second internal C-domain while BenY-C is the CT domain, as depicted in Figure 2D.
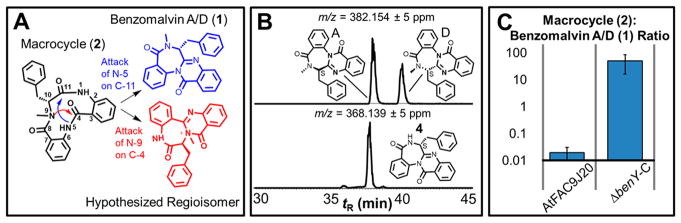
Figure 3.
Transannulation is mediated by BenY-CT. (A) Benzodiazepine formation could occur through two different transannulation paths. The path leading to benzomalvin A/D (1) is colored blue. The path leading to an undetected and unreported alternate regioisomer with a quaternary amide is colored red. (B) The top trace shows two chromatographic peaks in the extracted ion chromatogram for m/z 382.1547, corresponding to the atropisomeric pair of benzomalvin A and benzomalvin D, which are in conformational equilibrium and are collectively called benzomalvin A/D (1). The bottom trace shows the extracted ion chromatogram for desmethyl benzomalvin (4), which shows only one major chromatographic peak. Benzomalvin regioisomerism is expected to lead to four peaks and two peaks of similar abundance in the top and bottom traces, respectively, suggesting transannulation occurs with regioselectivity, regardless of methylation status. (C) The compound 2:compound 1 ratio is >100-fold increased by deletion of benY-CT, suggesting BenY-CT catalyzes transannulation of 2 to form 1. The p value for the difference between AtFAC9J20 and ΔbenY-C was calculated by a Student’s t test and equals 0.004, indicating a statistically significant difference.
The anthranilate-containing benzodiazepine natural product asperlicin C differs in structure from benzomalvin A/D only in its lack of N-methylation and the replacement of the Phe-derived phenyl ring with a tryptophan-derived indole. Asperlicin C is one of two equally abundant products of the fungal NRPS enzyme AspA, where the other product represents an alternative regioisomer, asperlicin D. Asperlicins C and D have both been shown to be formed through a non-enzymatic and non-regioselective transannulation reaction; however, despite several studies of the structure of benzomalvins, only one regioisomer (from the N-5 to C-11 transannulation reaction) of the benzomalvins has ever been reported (Figure 3A).
N-Methylation is an important structural feature of the benzomalvins and has been proposed to be the source of benzomalvin atropisomerism, by providing sufficient steric hindrance to slow the ring pucker conformational transition enough that the atropisomers can be chromatographically resolved.17 We hypothesized that this same N-methylation might also be responsible for benzomalvin regiospecificity, because the unobserved regioisomer would contain a disfavored quaternary amide. Previously, we reported that desmethyl benzomalvin (4) is produced as a side product of benzomalvin biosynthesis. To test the role of N-methylation in benzomalvin regiocontrol, we used LC–MS to compare the extracted ion chromatograms for benzomalvin A, benzomalvin D, and 4 in Figure 3B. If N-methylation is responsible for regiocontrol, we would expect to observe a mixture of regioisomers when the methyl group is absent. Interestingly, the extracted ion chromatogram of 4 displays only one predominant peak.
There are various possible explanations for the observation of only one desmethyl benzomalvin peak. One possibility is that two regioisomers with the same molecular formula as 4 are present but are not chromatographically resolved under the experimental conditions. However, the chemically related asperlicin regioisomers are reported to be well-resolved from one another under similar chromatographic conditions. Another possible interpretation of the current data is that multiple regioisomers of benzomalvin are present, but one predominates and is represented by the most abundant peak. The final interpretation is that there is only one regioisomer, corresponding to 4; this analysis supports the role of N-methylation in atropisomerism but indicates that regiocontrol is achieved in a manner independent of N-methylation status.
While the presence of an undetected regioisomer cannot be ruled out, we sought to evaluate alternate sources of regiocontrol, including the possibility that regiocontrol could be enzymatic. We hypothesized that BenY-CT might have the appropriate catalytic machinery to catalyze such a transannulation reaction itself and would be an ideal candidate for benzodiazepine synthase activity because it would already come into contact with 2, the substrate for such a reaction. To test this hypothesis, we compared the ratio of the macrocyclic benzodiazepine precursor (2) to benzomalvin A/D (1) in extracts both from AtFAC9J20 containing the full benzomalvin biosynthetic gene cluster and from AtFAC9J20-ΔbenY-CT (Figure 3C). If the conversion of 2 to 1 is non-enzymatic, then the presence or absence of BenY-CT should not have an effect on the 2:1 ratio. However, if the process is catalyzed by BenY-CT, then the 2:1 ratio might be significantly increased if non-enzymatic cyclization and release of 2 from the NRPS machinery occur at a rate similar to or faster than the rates of transannulation of 2 and formation of 1. Figure 3C shows that upon deletion of benY-CT, the 2:1 ratio is increased by ~100-fold, suggesting that BenY-CT either directly catalyzes transannulation or recruits another enzyme present in both the native A. terreus strain and the heterologous host A. nidulans. This finding strongly suggests the existence of a benzodiazepine synthase activity and marks BenY as a candidate to be the first known benzodiazepine synthase. The overall pathway of benzomalvin A/D biosynthesis is shown in Figure 4. Because benzodiazepines represent a valuable class of psychoactive drugs,9,13 the discovery of a benzodiazepine synthase may lead to the discovery of new valuable molecules through genome mining, or new semisynthetic approaches for benzodiazepine production. Future studies demonstrating in vitro reconstitution of the isolated enzyme will be needed to confirm this finding and to focus on the specific catalytic machinery involved in transannulation.
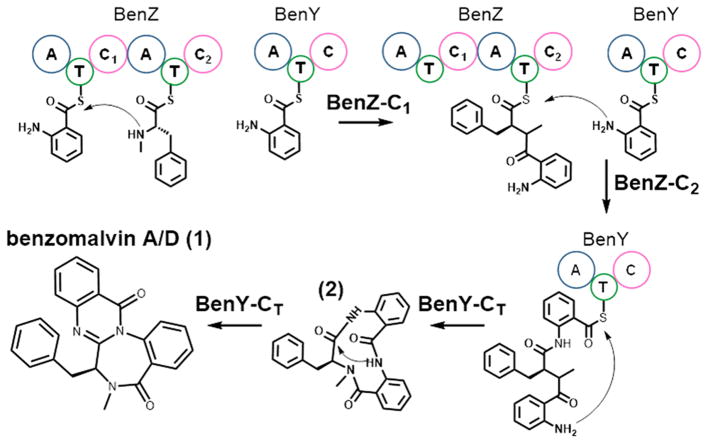
Figure 4.
Biosynthesis of benzomalvin A/D. Biosynthesis proceeds through formation of the Anth-NmPhe dipeptide covalently bound via a thioester bond to the second T-domain of BenZ. Then, BenZ-C2 catalyzes formation of the Anth-NmPhe-Anth tripeptide covalently bound to BenY-T. BenY-CT then catalyzes cyclization and cleavage of the thioester bond, leading to transannulation and production of benzomalvin A/D.
CONCLUSIONS
Herein, the application of targeted domain deletions within a FAC-encoded biosynthetic gene cluster enabled us to assign specific biosynthetic activities to individual NRPS domains, leading to identification of BenY-C as the enzyme domain responsible for cyclization and release of the benzomalvin precursor (2), as well as its transannulation to form 1. We also assigned specific roles to BenZ-C1 and BenZ-C2, as well as their corresponding adenylation domains. Future work will seek to further elucidate the details of benzomalvin biosynthesis, including analysis of the enzyme mechanism for transannulation and determination of the enzymes involved in formation of the additional benzomalvin family members benzomalvin B, C, and E. Because bioinformatic analyses cannot currently differentiate between the cyclizing BenY-C domain and the noncyclizing BenZ-C2 domain, this study may also assist the development of a bioinformatic signature to differentiate such domains in the future. More broadly, molecular genetic approaches are attractive for dissection of natural product biosynthesis,18,19 but the availability of genetic tools for performing such analyses is dependent on the fungal species under study; a vast majority of fungi lack suitable genetic tools. This work has shown the feasibility of using FACs to analyze precisely fungal metabolite biosynthesis through recombineering in E. coli, theoretically enabling application to any filamentous fungal species. In the future, the approach demonstrated here of in-cell dissection of biosynthesis should greatly aid the study of fungal specialized metabolism and the discovery of new fungal enzyme activities.
Supplementary Material
Acknowledgments
Funding
The authors thank the National Institutes of Health for funding to N.L.K., J.W.B., and C.C.W. (SBIR Grant R44AI118086 from the National Institute of Allergy and Infectious Diseases), to N.L.K. (Grant R01AT009143), to N.P.K. (Grant R01AI065728), to G.P.M. (Grant T32GM105538), and to M.T.R. (Grant T32GM008449).
Footnotes
Notes
The authors declare the following competing financial interest(s): C.C.W., R.Y., M.N.I., C.C., M.S., and E.W. are employees of Intact Genomics that sells the unbiased Random Shear BAC and FAC libraries and services for genome discovery and DNA research, as well as BAC cloning, DNA end repairing, E. coli competent cells, and other enzyme kits for DNA and protein research.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.8b00076.
Figures S1 and S2 and Tables S1 and S2 (PDF)
References
- 1.Blackwell M. The fungi: 1, 2, 3··· 5.1 million species? Am J Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 2.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, Wymore F, Wortman JR, Sherlock G. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han X, Chakrabortti A, Zhu J, Liang ZX, Li J. Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae. BMC Genomics. 2016;17:633. doi: 10.1186/s12864-016-2974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discovery. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 6.Dias DA, Urban S, Roessner U. A Historical Overview of Natural Products in Drug Discovery. Metabolites. 2012;2:303–336. doi: 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clevenger KD, Bok JW, Ye R, Miley GP, Verdan MH, Velk T, Chen C, Yang K, Robey MT, Gao P, Lamprecht M, Thomas PM, Islam MN, Palmer JM, Wu CC, Keller NP, Kelleher NL. A scalable platform to identify fungal secondary metabolites and their gene clusters. Nat Chem Biol. 2017;13:895–901. doi: 10.1038/nchembio.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bok JW, Ye R, Clevenger KD, Mead D, Wagner M, Krerowicz A, Albright JC, Goering AW, Thomas PM, Kelleher NL, Keller NP, Wu CC. Fungal artificial chromosomes for mining of the fungal secondary metabolome. BMC Genomics. 2015;16:343. doi: 10.1186/s12864-015-1561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun HH, Barrow CJ, Sedlock DM, Gillum AM, Cooper R. Benzomalvins, new substance P inhibitors from a Penicillium sp. J Antibiot. 1994;47:515–522. doi: 10.7164/antibiotics.47.515. [DOI] [PubMed] [Google Scholar]
- 10.Jang JP, Jang JH, Soung NK, Kim HM, Jeong SJ, Asami Y, Shin KS, Kim MR, Oh H, Kim BY, Ahn JS. Benzomalvin E, an indoleamine 2,3-dioxygenase inhibitor isolated from Penicillium sp. FN070315. J Antibiot. 2012;65:215–217. doi: 10.1038/ja.2011.141. [DOI] [PubMed] [Google Scholar]
- 11.Ames BD, Walsh CT. Anthranilate-activating modules from fungal nonribosomal peptide assembly lines. Biochemistry. 2010;49:3351–3365. doi: 10.1021/bi100198y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 13.Haynes SW, Gao X, Tang Y, Walsh CT. Assembly of asperlicin peptidyl alkaloids from anthranilate and tryptophan: a two-enzyme pathway generates heptacyclic scaffold complexity in asperlicin E. J Am Chem Soc. 2012;134:17444–17447. doi: 10.1021/ja308371z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 15.Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT, Tang Y. Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat Chem Biol. 2012;8:823–830. doi: 10.1038/nchembio.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Liu N, Cacho RA, Gong Z, Liu Z, Qin W, Tang C, Tang Y, Zhou J. Structural basis of nonribosomal peptide macrocyclization in fungi. Nat Chem Biol. 2016;12:1001–1003. doi: 10.1038/nchembio.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun HH, Barrow CJ, Cooper R. Benzomalvin D, a New 1,4-Benzodiazepine Atropisomer. J Nat Prod. 1995;58:1575–1580. [Google Scholar]
- 18.Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CC. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henke MT, Soukup AA, Goering AW, McClure RA, Thomson RJ, Keller NP, Kelleher NL. New Aspercryptins, Lipopeptide Natural Products, Revealed by HDAC Inhibition in Aspergillus nidulans. ACS Chem Biol. 2016;11:2117–2123. doi: 10.1021/acschembio.6b00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 21.Muyrers JP, Zhang Y, Stewart AF. Techniques: Recombinogenic engineering–new options for cloning and manipulating DNA. Trends Biochem Sci. 2001;26:325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 22.Johnson SJ, Wade-Martins R. A BACwards glance at neurodegeneration: molecular insights into disease from LRRK2, SNCA and MAPT BAC-transgenic mice. Biochem Soc Trans. 2011;39:862–867. doi: 10.1042/BST0390862. [DOI] [PubMed] [Google Scholar]
- 23.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Düsterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJN, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kötter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nützmann HW, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gómez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage AA, Caffrey P, Cheng YQ, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, van der Donk WA, Dorrestein PC, Doyle S, Driessen AJM, Ehling-Schulz M, Entian KD, Fischbach MA, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Höfte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon HJ, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Méndez C, Metsä-Ketelä M, Micklefield J, Mitchell DA, Moore BS, Moreira LM, Müller R, Neilan BA, Nett M, Nielsen J, O’Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet JL, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas JA, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Süssmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, Glöckner FO. Minimum Information about a Biosynthetic Gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.